GeneNet database: a technology for a formalized description of gene networks
Ananko E.A., Kolpakov F.A., *Kolchanov N.A.
Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia
e-mail: genenet@bionet.nsc.ru
Keywords: gene networks; database; signal transduction pathways; visualization
Resume
Motivation:
Information regarding peculiarities of gene networks organization and mechanisms of their functioning is rapidly increasing. For integration of this sort of knowledge, it is necessary to develop effective computer technology enabling to describe and visualize all integrity of elementary structures and processes in gene networks in both pro- and eukaryotes.
Results:
The computer technology for a formalized hierarchical description of a gene network has been developed. The database on gene networks (GeneNet) and the software for its automated visualization have been worked out. To provide rapid data accumulation, the Java graphic interface for the data input through the Internet is developed. Currently, GeneNet database accumulates descriptions of 18 gene networks subdivided between six sections.
Availability:
The GeneNet is available at
http://wwwmgs.bionet.nsc.ru/mgs/gnw/genenet/Introduction
Gene network is an ensemble of coordinately expressed genes controlling vital functions [Kolpakov et al., 1998]. Regulation of the gene network operating is not restricted to the level of transcription, but it may be carried out at the levels of translation [Pyronnet et al., 1996; Buss and Stepanek, 1993], splicing [Yao et al., 1996; Pyronnet et al., 1996; Nandabalan and Roeder, 1995], posttranslational protein degradation [Hochstrasser, 1996], active membrane transport [Weissmuller and Bisch, 1993], and other processes.
The pioneer theoretical investigation of gene networks is dating back to 60-ties. These studies were devoted to consideration of general regularities of molecular-genetic system regulation in procaryotes [Ratner, 1966] and to description of gene network dynamics within the frames of the simplest logical models [Kauffman, 1969]. Later on, for the studying of gene networks dynamics, the approaches based on application of differential equations and stochastic models were suggested [Savageau, 1985; Thomas et al., 1995; McAdams and Arkin, 1997]. For integration of heterogeneous experimental information and its accumulation in databases, an effective computer technology is necessary, which permits to describe all variability of elementary structures and processes occurring in gene networks of pro- and eukaryotes within the frames of a unified approach. Moreover, it is desirable to produce an automated visualization of the gene network structure based on the formalized information stored in the database.
In the GeneNet system presented [Kolpakov et al., 1998; Kolpakov and Ananko, 1999], we have applied an original computer technology. It permits to describe any elementary structure and event occurring in pro- and eukaryotic gene networks at different hierarchical levels of organization, i.e., molecular, cellular, and referring to the whole organism levels.
Methods and algorithms
Experimental data from original papers are formalized and collected in the GeneNet database. An object-oriented approach [Booch, 1991] was employed as a basis for describing the gene network structure. For each gene network, several obligatory types of structural and functional components are marked out: 1) a gene ensemble interacting when certain biological functions are performed (the core of a gene network); 2) proteins encoded by these genes and respected for structural, transport, catalytic, regulatory, and other functions; 3) signal transduction pathways providing gene activation in response to external signals; 4) a set of positive and negative feedbacks stabilizing the parameters of the gene network (autoregulation) or providing a transition to a new functional state; 5) nonproteinaceous substances such as signal molecules, energetic cell components, metabolites, etc. [Kolpakov et al., 1998].
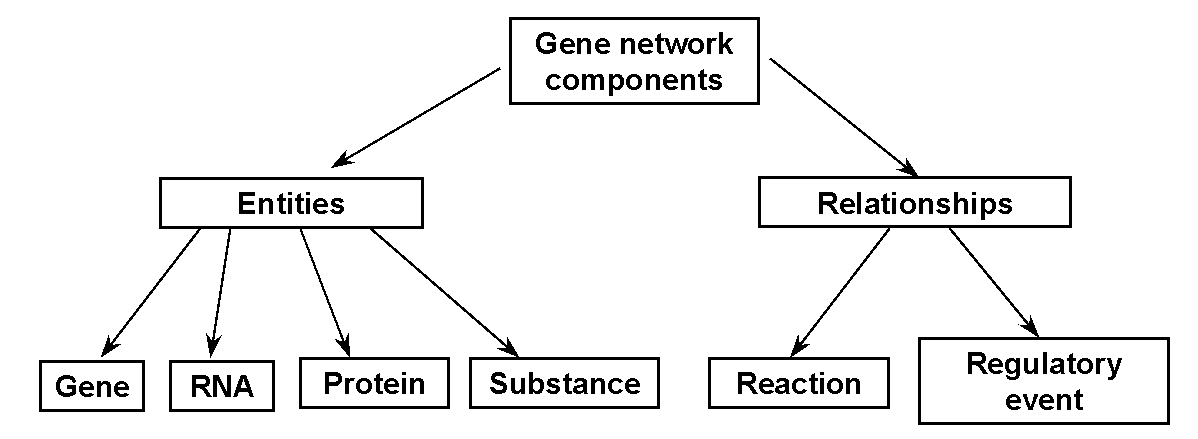
All the gene network components are divided into the Entities (any material objects) and Relationships between the gene network components (Fig.1).

The entities are subdivided into 4 classes: 1) Protein or protein complex; 2) Gene; 3) RNA; 4) nonproteinaceous Substance. Instances of each class are described in a separate table in the GeneNet database The components of a gene network are scattered throughout cell compartments, cells, tissues and organs [Kolpakov et al., 1998].
Two types of relationships between the entities are considered: Reaction, that is, formation of a new entity or acquisition of a new property by the entity, and Regulatory event, that is, the effect of an entity onto a certain reaction.
The formalized description of elementary events is based on application of several informative line codes. Example is given below:
ID <gene>Hs:OAS^nucleus ->
<protein>Hs:OAS^cytoplasm
DT 17.5.1999; Ananko E.; created.
EF indirect
RF Wathelet M. et al., 1986
//
It means that the protein encoded by the human oligoadenylate synthase (OAS) gene is expressed in a cytoplasm (line code ID). The relationships is indirect (line code EF), because the intermediate stages such as transcription, processing, splicing, etc. are missing.
The phosphorylation initiated by the interferon receptor II (IFNR-II) of the human protein kinase Jak1 in the cell cytoplasm is described as follows:
ID
<protein>Hs:IFNR-II^cytoplasm ->> <protein>Hs:Jak1^cytoplasm ->
<protein>Hs:Jak1-p^cytoplasm
DT 17.5.1999; Ananko E.; created.
AT switch on
EF indirect
RF Silvennoinen, O. et al., 1993
//
Similarly, any other elementary event may be described in the terms of GeneNet, for instance, mRNA translation, enzyme catalysis, multimerization, etc.
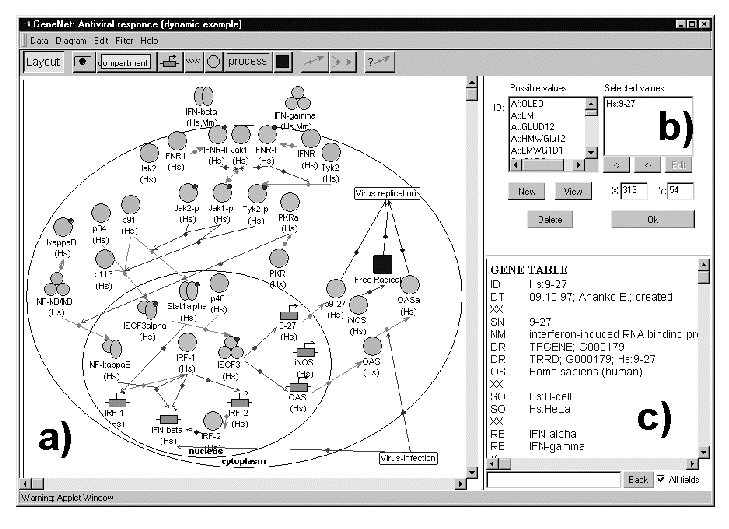
To provide rapid data accumulation in the database, the interface for data input through the Internet was developed, which enables the user to add novel objects into the database, establish relationships between them, and to transform automatically the information into the GeneNet language (Fig. 2).
The chief merit of the technology developed is a possibility to make automated visualization of the gene network diagram. The formalized data on the gene network stored in the GeneNet database are processed by the special Java-program (GeneNet viewer) and then the data are shown up to the user as a graphical diagram (Fig. 2).
Implementation and results
All the images at the diagram are interactive, that is, if a user clicks the image, the textual description of the corresponding entry in the GeneNet database is displayed in a special text window (Fig. 2c). The text window contains a formatted text with hypertext references to other
 Figure
2. GeneNet graphic user interface for data input through the Internet Figure
2. GeneNet graphic user interface for data input through the Interneta) fragment of a gene network regulating anti-viral response; b) interactive data input window; c) formalized description of an object (the human gene 9-27). |
databases (EMBL, SWISS-PROT, TRRD, TRANSFAC, EPD, MEDLINE) and links with other GeneNet tables (Fig. 2).
Informational content of GeneNet database is given in Table 1. By the 1st of April, 2000, 18 gene networks referring to 6 sections are described in the database.
Table 1. Informational content of GeneNet database (by the 1st of April, 2000)
| Thematic section | Authors | Number of gene networks | Number of components | ||
| genes | proteins | interactions | |||
| Lipid metabolism | Ignatieva E.V. | 2 | 16 | 22 | 86 |
| Endocrine regulation | Busygina T.V. Ignatieva E.V. Logvinenko N.S. Suslov V.V. |
4 | 31 | 95 | 187 |
| Erythropoiesis | Podkolodnaya Î.À. | 1 | 32 | 51 | 87 |
| Anti-viral response | Ananko Å.À. | 1 | 12 | 51 | 65 |
| Development of seeds in plants | Goryachkovsky T.N. Aksenovich A.V. | 9 | 45 | 97 | 390 |
| Heat shock | Stepanenko I.L. | 1 | 4 | 16 | 34 |
| Ttotal: | 16 | 128 | 289 | 765 | |
Discussion and conclusions
At present, several databases are known describing different aspects of gene network organization, e.g., CSNDB (Cell Signaling Networks Database) [Igarashi and Kaminuma T, 1997] contains an information about signal transduction mechanisms in the human cells; BRITE (Biomolecular Reaction pathways for Information Transfer and Expression) [Hopkins, 1995] accumulates the data on the cell cycle genes as well as the schemes for the pathways controlling early development in Drosophila; GeNet (Gene Networks database) [Serov, 1998] describes gene networks of Drosophila, Nematode caenorhabditis and Echinus esculenta; KEGG (Kyoto Encyclopedia of Genes and Genomes) [Kanehisa and Goto, 2000] stores the schemes of signal transduction pathways, genome maps, information about the genes; SPAD (Signaling Networks Database) (http://www.grt.kyushu-u.ac.jp/spad/) contains the structure-functional data on the mechanisms of signal transduction; EcoCyc [Karp et al., 2000] describes the metabolic pathways. However, none of these databases provides the solving of the whole complex of tasks necessary for a gene network effective studying, which demands analysis of the large bulk of heterogeneous experimental data.
The experience of this sort databases development makes clear the necessity of creating such universal computer technology that may describe any elementary structures, events, and processes significant for gene network operating. The computer technology GeneNet suggested [Kolpakov et al., 1998; Kolpakov and Ananko, 1999] is the way to the rapid accumulation of experimental data on structure-functional gene network organization, together with capacities for systematization and computer and logical analysis of this information.
Further development of the GeneNet database will proceed according to the following three directions: 1) improving of a gene network description with accounting of its hierarchical organization and spatial distribution; 2) development of approaches for mathematical modeling of gene network dynamics on the base of information stored in the GeneNet database.
Acknowledgments
The work was supported by the Russian Foundation for Basic Research (grants Nos. 98-04-49479, 98-07-91078, 99-07-90203, 00-07-90337, 00-04-49229, 00-04-49255), Russian Human Genome Program, Ministry of Science and Technology of Russian Federation, Integrated Program of the SB RAS. The authors are grateful to E.V. Ignatieva, O.A. Podkolodnaya, I.L. Stepanenko, T.N. Goryachkovsky, T.V. Busygina, A.V. Aksenovich, and V.V. Suslov for the database filling and helpful discussions; N.L.Podkolodny, and D.A. Grigorovich for SRS and software support; and G.V. Orlova for translation of the paper into English.
References