A.E. Kel', N.A. Kolchanov, 0.V. Kel', A.G. Romashchenko, E.A. Anan'ko, E.V. Ignat’eva, T.I. Merkulova, 0.A. Podkolodnaya, I.L. Stepanenko, A.V. Kochetov, F.A. Kolpakov, N.L. Podkolodnyi, and A.N. Naumochkin
Institute of Cytology and Genetics, Siberian Division, Russian Academy of Sciences, Novosibirsk, 630090 Russia
E-mail: kel@bionet.nsc.ru
Received October 31,1996
Abstract—The format of the TRRD complies with the model of hierarchical organization of transcription regulatory regions of eukaryotic genes. The current version of TRRD covers more than 300 genes, with several large sections concerning different important systems of functionally linked genes. Each such section contains formalized information on the structure of regulatory regions in a specially designed format, and nonformalized information in textual and graphic form. Integration is achieved with the WWW hypertext tools. We describe a relational model of the database and ways for its integration with other databases, first of all TRANSFAC and COMPEL. TRRD is accessible at http//www.bionet.nsc.ru/TRRD/
Key words: eukaryotic gene transcription regulation, databases
Studies on the regulation of eukaryotic gene expression are one of the central activities in molecular biology. Rapid accumulation of experimental data in this field necessitates development of computer databases for their systematization, storage, and retrieval.
Creation of such databases is a quite complicated task, which is determined by the complex multistage mechanisms involved in gene expression. Along with transcription, it includes pre-mRNA capping, splicing, 3'-terminal processing/polyadenylation, RNA transport across nuclear membranes, and mRNA translation into a polypeptide. The intensity of each process is controlled by different mechanisms. However, transcription is the key event initiating the above chain, and this explains the special interest in investigating its regulation.
The eukaryotic transcription machinery is under complex control acting differentially on particular genes depending on the cell cycle stage [1-3], cell or tissue type [4-9], stage of ontogeny [10] and organismic functional status [II, 12], as well as environmental conditions [13,14].
Regulation involves a vast variety of processes taking place at the levels of separate genes, chromosomes, cell nuclei, cells, tissues, and the entire organism (see introductory paper [15]). Notwithstanding, the central part therein is played by genomic regulatory sequences. Crucial for initiating transcription of any gene read by RNA polymerase II is the core promoter: a short stretch of DNA located at the transcription start and containing a TATA box and an Inr element [16]. The core promoter is the assembly site for the basal transcription complex, which, along with RNA polymerase II, includes universal transcription factors such as TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH [17-19].
The required intensity of transcription of a particular gene in particular conditions is set by the gene-specific transcription complex. The latter is formed owing to (i) interaction of specific nuclear transcription factors with (cis-regulatory [20] and composite elements [21] in enhancers and silencers, and (ii) interaction of such DNA-protein complex with the basal transcription complex [22-26]. Essential for adjusting the transcription are the regulatory elements in the 5'-flanking regions, introns [27, 28], and at the 3' termini of eukaryotic gene [29].
Our goal was to build a database on the structure and function of the above-mentioned regulatory regions in genomic DNA: TRRD (Transcription Regulatory Region Database) based on the model [30] that accounts for the diversity of transcription-controlling elements, their modular structure, and hierarchy essential for their functional manifestation.
The current TRRD version has several large sections devoted to different systems of functionally coupled genes, such as interferon-inducible genes, lipid metabolism genes, cell cycle control genes, etc. Each such section contains formalized information on the structure of regulatory regions in a specially designed format, and nonformalized information in textual and graphic form. Integration is achieved with the WWW hypertext tools.
Here we describe the TRRD format [14], present the formalized description of a number of regulatory regions with qualitatively different organizational features, and consider the ways for its integration with other databases, first of all TRANSFAC [31] and COMPEL [21]. The TRRD sections on gene functional families are described in detail in this issue [32-35].
One of the key tasks in designing the TRRD format was to develop a formalized description for experimental data on the structural and functional organization of genomic DNA regions involved in transcription regulation. The format had to be flexible enough to admit new data without essential changes in the previous information and to describe the results obtained with the most popular techniques, to reflect the established concepts on the structure and function of the gene regulatory regions and to allow integration with other databases.
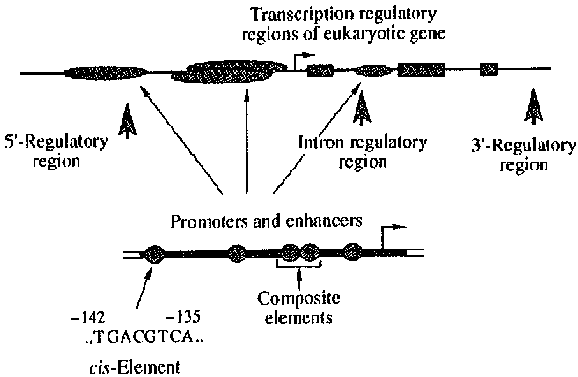
In accordance with the above, we chose a format capable of describing the modular structure of transcription regulatory regions and the hierarchy of their constituent units (Fig. 1):

Fig. 1. Hierarchical organization of transcription regulatory regions of eukaryotic genes.
This scheme corresponds to the general model of eukaryotic gene transcription regulatory regions; however, separate elements thereof may be absent owing to specific regulation of particular genes or because of the lack of experimental data.
TRRD includes only experimentally confirmed information. A database entry corresponds to regulatory regions of a single gene, and is updated as new evidence appears. Each entry is a set of fields arranged in accord with the above modular hierarchy. Any field may contain reference to the original paper, another database, or other information source. Complete references to publications are given at the end of each entry.
Let us consider the main database entry fields, with human glycoprotein hormone (chorionic gonadotropin) a -subunit (a -GH) gene as an example.
ID: identifier consisting of abbreviated species and gene names, or abbreviated virus name for single-transcript viruses.
OS: full species name (English and/or Latin).
SN: short gene name.
NG: full name of gene or eukaryotic single-transcript virus (synonyms parenthesized).
CG: gene classification according to the EPD system [40]; e.g., (a -GH belongs to chromosomal genes of vertebrates (6.1); subclass of hormones, growth factors, and regulatory proteins (6.1.5); family of placental and hypophysial hormones (6.1.5.3); subfamily of glycoprotein hormones (6.1.5.3.1).
RE: qualitative description of the specific gene regulatory features in four main groups: (1) cell cycle stage at which gene transcription is activated or suppressed; (2) ontogenetic stage at which the gene is expressed (e.g., embryo, fetus, mature organism); (3) organ, tissue, and cell type exhibiting activated or suppressed transcription; (4) inducing and repressing factors and stimuli (e.g., heat shock, starvation, chemicals, hormones, interleukins, growth factors, vitamins, etc.); changes in transcription are marked with plus or minus signs. Thus a fragment below shows that the (a -GH gene is tissue-specific, its expression is suppressed by steroid and thyroid hormones, and enhanced by cAMP:
RE 3(+): placenta, gonadotropes of the anterior pituitary [Bokar J.A. et al., 1989]
RE 4(-): steroid hormones [Akerblom et al., 1988, Clay et al., 1993]
RE 4(-): thyroid hormone [Chatterjee et al., 1989]
RE 4(+): cAMP [Bokar J.A. et al., 1989]
KW: key words essential for grouping genes with similar structural or functional characteristics (e.g., involvement of protein products in a common metabolic pathway, or close gene location on the chromosome).
CH: chromosomal assignment or gene location if known.
A set of eleven fields describes each of the transcription factor binding sites found in the gene. Sites are listed from the 5' to the 3' end.
AN: site accession number.
NM: site brief and full names (synonyms parenthesized).
NS: reference to the given site in TRANSFAC.
TF: brief name of transcription factor, full name, source species.
In cases when the gene regions has been shown to interact with several factors belonging to the same functional family and acting in the same direction, these factors are listed with plus signs.
AT: increase or decrease in the transcription rate upon factor binding to the DNA site.
SQ: nucleotide sequence of the fragment corresponding to the consensus of the given type of site.
PQ: positions of the beginning and the end of SQ relative to the position given in PR (vide infra).
SF: sequence of the site region as determined by footprinting.
PF: start and end positions of SF.
The two sequence descriptions are necessary because in most cases the footprinting region does not coincide with the consensus site.
BF: beginning of the site in the EMBL sequence.
AG: listing of cell lines and types of experiment in numerical notation: (1) footprinting, methylation, and gel retardation: locating the site and identifying the transcription factor; (2) genetic engineering of vectors with the regulatory zone under study and artificial regulatory zones in preset context: functional assessment; (3) combination of the above with site-directed mutagenesis: revealing the functional role of separate nucleotides; (4) phylogenetic comparisons: approximate location of the functional region.
Further, there are three commentary fields: HM describing homologous sites, EF specifying the relative affinity of the given site to the transcription factor, and KK with other comments.
A fragment of the description of one site in the (a -GH gene is given below. Fuller examples can be found in the Appendix.
AN 176
NM GRE(I); glucocorticoid responsive element
NS R00591
TF GR; glucocorticoid receptor [Akerblom et al., 1988]
AT decrease
SQ ACATCAAATTCACGT
PQ -151 to -137 (C)
BF HSGPHA:185
AG 1.1.5, 4.2, 6.1.l+, 7.1 [Akerblom et al., 1988]
CE: description of composite elements, with the COMPEL index, start and end positions, listing of constituent sites, and references:
CE GATA$UNCL_001; C00026; -172 to –151; 172, 1067 [Steger D.J. et al., 1994]
PR: description of promoter, enhancer, or silencer, specifying: the name of regulatory unit; the reference point (ST, transcription start; SR, translation start; SS, sequence start) and location of promoter, enhancer, or silencer; numbers of all sites (as in AN) related to the given units. For the (a -GH gene, this appears as follows:
PR placenta-specific enhancer; ST; -169 to -100; 172, 1067, 173, 174, 175
PR [Bokar J.A. et al., 1989, Andersen et al., 1990]
XX
PR steroid-responsive region; ST; -160 to –60; 176, 177, 178
PR [Akerblom et al., 1988]
XX
PR proximal promoter; ST; -90 to+l; 179, 180, 181
PR [Bokar et al., 1989, Chatterjee et al., 1989]
Sand means that the 5' flank contains a promoter and two enhancers; interestingly, the three elements partly overlap, which is typical of eukaryotic genes and can be described in the proposed format.
RG: gene part containing the regulatory region (5' or 3' flank, exon, intron; LTR for eukaryotic viruses; etc.).
MP: mutual arrangement of promoters, enhancers, and silencers in all gene regulatory regions. Such a map is specified if there are several transcription starts depending on the promoter, or if different regulatory regions have been studied independently and only their approximate relative position is known. For instance, this field in the case of the 6-bisphosphatase gene (G000784, PFK-2/FBPase-2)
MP SS 1808 ST1 < ST2
indicates that the F promoter begins 1808 nt upstream of the transcription start from the M promoter which itself is some distance upstream of the L promoter transcription start.
The description of all known regulatory regions of the given gene is followed by comments pertaining to the gene as a whole (synergism between remote sites, functional interaction of regulatory blocks, protein-protein interaction between transcription factors and with elements of the basal complex) and other info at the referee's discretion.
Each entry ends with a full list of references (AUthor(s), TItle, YeaR, SOurce, VoLume, PaGes),
The full a -GH gene description in this format is given in the Appendix, and shows that its transcription is controlled by a proximal promoter, two enhancers, two composite elements, and 11 transcription factor-binding sites.
The TRRD data can be visualized with the MGL system (see special paper in this issue [41]), which displays a graphic map of the gene regulatory array. Visual analysis of the TRRD testifies to the qualitative diversity of eukaryotic transcription regulation.
The MGL system can represent the TRRD data as a regulatory map of an entire gene as well as those of its separate regions in more detail.
The a -GH gene (Fig. 2) has a quite complex structure of the 5' regulatory region. Indeed, a relatively short stretch (some 150 bp) directly upstream of the transcription start harbors a proximal promoter and two enhancers (one tissue-specific and the other steroid-responsive), with two composite elements and 11 binding sites for different transcription factors. This is a vivid example of the modular structure of transcription-controlling regions. It also shows that the 5' regulatory regions may have an exceptionally high functional coding density.
Fig. 2. Fragment of a regulatory map of the a -GH gene (G000271 in TRRD) with two enhancers (I, placenta- and II, steroid-specific) and promoter (III); arrows mark two composite elements.
Figure 3 depicts fragments of the regulatory map for the human P-globin gene. There are four large blocks. The first one is a promoter occupying positions -220 to +20 (Fig. 3c); in addition to the TATA and CAAT boxes, it contains numerous transcription factor-binding sites, including two for the erythroid-specific factor GATA-I (NF-EI). The second and the third blocks are silencers, -610 to -490 (Fig. 3a) and -338 to -233 (Fig. 3b); both suppress the gene transcription in K562 cells, the latter one being more effective [42]. The fourth block is an enhancer more than 2000 bp downstream of the transcription start (Fig. 3d), which modulates tissue-specific transcription and is ontogenetically important [43].
This gene also exhibits an interesting feature of transcription regulatory regions: multiply repeated sites, such as the NF-E1 (GATA-1) found both in the promoter and in the remote 3' enhancer parts.
Still another regulatory element of this gene is the locus-controlling region (LCR) more than 60 kbp upstream of the transcription start. It consists of four hypersensitive sites, two of which are described in the database; their regulatory maps are shown in Fig. 4. Very remote enhancers are found for many eukaryotic genes. As follows from Figs. 3 and 4, LCR, promoters, and enhancers have a number of identical sites such as GATA-I (NF-E1) and CAC boxes [35], which is indicative of certain similar steps in the functioning of promoters and LCR units of a higher rank.
Fig. 3. Regulatory map of the human b -globin gene (G000215).
Fig. 4. Regulatory map of enhancer hypersensitive sites in the LCR of the human b -globin gene cluster (G000328).
The TRRD version 3.3 describes 355 genes, 493 promoters, enhancers, and silencers; and 1740 binding sites for transcription factors. This information originates from more than a thousand publications. The database represents genes of human (148 entries), mouse (98), rat (58), chicken (22), viruses (II), frog (6), rabbit (6), and other organisms (7).
As indicated above, TRRD considers four main types of regulatory situations and accordingly groups the genes as (1) 43 cell cycle-dependent, (2) 87 onto-genetically dependent, (3) 277 tissue-specific, and (4) 259 responsive to external stimuli; some genes may have several types.
As can be seen in Table I, the largest number of tissue-specific genes has been found in liver, blood cells, and muscle.
Table 1. Tissue-specific genes presented in TRRD
Tissue |
Number |
of genes |
inducible |
repressible |
|
| Liver | 98 |
12 |
| Kidney | 13 |
11 |
| Pancreas | 1 |
5 |
| Placenta | 10 |
1 |
| Brain | 5 |
12 |
| Blood cells | 42 |
5 |
| Muscle | 23 |
4 |
| Spleen | 6 |
2 |
| Intestine | 8 |
1 |
Our relational model (Fig. 5) uses FoxPro for Windows and specially designed programs for data conversion from the TRRD to the relational database format.
Fig. 5. Flowsheet of the TRRD relational model.
The model comprises 24 tables (15 with data and 9 liaison ones); the main data tables are listed in Table 2; all tables are interlinked in one-to-one and one-to-many modes.
Table 2. Relational tables in TRRD
| Name | Number of entries |
Description |
| T_GENE.DBF | 355 |
Gene table |
| T_KW.DBF | 1056 |
Key words |
| T_RE.DBF | 1139 |
Gene regulation table |
| T_PROM.DBF | 493 |
Promoters and enhancers |
| T_CE.DBF | 74 |
Composite elements |
| T_SITE.DBF | 1740 |
Binding sites |
| T_FACTOR.DBF | 1333 |
Transcription factors |
| T_AG.DBF | 2844 |
Identification methods |
| LITER.DB | 1169 |
References |
| T_SEQ | 2254 |
Nucleotide sequences |
Figure 5 also shows the linking of TRRD with EMBL, TRANSFAC, and COMPEL. On this basis one can make complex queries on gene sequences, transcription factors, composite elements, etc.
Besides the above-mentioned visualization program, there is the S_TRRD program based on the relational representation, which offers a broad scope of procedures for data handling: filling the database, editing of the existing entries, complex search versions, etc. This program has a friendly multi-window interface with menus and an elaborate help system. It permits various operations with nucleotide sequences of regulatory regions, such as reading them from EMBL according to identifiers in the BF field, compiling samples of DNA sequences that include transcription factor-binding sites, composite elements, and promoter regions (such samples are necessary for computer analysis of these sites).
The WWW tools were used to create a hypertext system WWWTRRD, which gives Internet users access to the TRRD information, integrates the different TRRD parts and sections, and links TRRD with other databases.
TRRD has several large sections devoted to description of the functionally important gene systems presented in Table 3. Thus the current version describes the main types of regulation known for eukaryotic cells: tissue-specific (e.g., glucocorticoid-responsive genes and lipid metabolism genes, many of which are expressed in liver, and muscle genes), regulation by inducers (glucocorticoid hormones and interferons), regulation in the cell cycle, and ontogenetic regulation (erythroid-specific genes).
Table 3. TRRD sections
| Gene systems | Number of genes in TRRD |
| Interferon-inducible | 60 |
| Lipid metabolism | 40 |
| Cell cycle control | 20 |
| Glucocorticoid-responsive | over 30 |
| Muscle-specific | 20 |
| Erythroid-specific | 33 |
Each TRRD section comprises several interlinked hypertext parts providing a description of the general characteristics of the gene group, formalized description of their regulatory regions, and other textual information, schemes and figures representing special features. Hypertext links among documents allows retrieval and viewing of any textual and graphic information. The WWW tools can extract formalized information for visualizing the gene regulatory maps.
The WWWTRRD system provides linking with EMBL, TRANSFAC, COMPEL, and other relevant databases.
We have created the TRRD to describe the structural and functional organization of the transcription regulatory regions of eukaryotic genes. The database provides formalized and hypertext (WWW tools) description of transcription regulation for more than 300 genes.
The most promising direction in further TRRD development is its integration with TRANSFAC, COMPEL, EPD, and other databases accumulating information of gene regulation.
TRRD is accessible via Internet at http://www.bionet.nsc.ru/trrd/
We are grateful to E. Wingender for fruitful discussion on database integration. This work was supported by the State program on Human Genome, Russian State Committee on Science and Technology, Russian Foundation for Basic Research (project nos. 13241 a and 12757a), DOE (OR00033-93CIS002), NATO, and the Siberian Division of the Russian Academy of Sciences.
APPENDIX
TRRD entry for human glycoprotein hormone a -subunit gene.
ID Hs:GHA
DT 25/10/95
AC 00056
GN G000271
CR Kel O.
OS human
SN alpha-GH
NG glycoprotein hormone alpha subunit gene (chorionic
NG gonadotropin alpha-subunit gene)
CG 6.1.5.3.1
XX
RE 3(+): placenta, gonadotropes of the anterior pituitary
RE [Bokar J.A. et al., 1989]
RE 4(-): steroid hormones [Akerblom et al., 1988, Clay et al., 1993]
RE 4(-): thyroid hormone [Chatterjee et al., 1989]
RE 4(+): cAMP [Bokar J.A. et al., 1989]
KW hormone, glycoprotein
XX
RG 5'region
XX
PR placenta-specific enhancer; ST; -169 to -100; 172, 1067, 173, 174, 175
PR [Bokar J.A. et al., 1989, Andersen et al., 1990]
XX
CE GATA$UNCL_001; C00026; -172 to -151; 172, 1067 [Steger D.J. et al., 1994]
XX
AN 172
NM URE (TSE); upstream regulatory element (tissue-specific element)
NS R00213, R00590, R02655
TF TSEB; trophoblast-specific element binding protein (in placenta cells)
TF [Steger D.J. et al., 1994]
AT increase
SQ AAGGGTTGAAACAAGA
PQ -169 to -154
BF HSGPHA:165
AG Placenta cells: 3.4, 6.1, 6.3 [Bokar J.A. et al., 1989]
AG Placenta cells: 1.1.1, 1.1.2, 3.6, 6.2, [Steger D.J. et al., 1994]
XX
CE ZIP$GATA_001; C00088; -156 to -135; 1067, 173 [Steger D.J. et al., 1994]
XX
AN 1067
NM GATA-binding site;
TF hGATA-2; + hGATA-3, in placenta cells; [Steger D.J. et al., 1994]
TF mGATA-2; + mGATA-4, in pituitary cells [Steger D.J. et al., 1994]
AT increase
SQ AGATAA
PQ -156 to -151
AG Placenta, pituitary cells: 1.1.1, 1.1.2, 3.4, 3.6, 6.2
AG [Steger et al., 1994]
XX
PR steroid-responsive region; ST; -160 to -60; 176, 177, 178
PR [Akerblom et al., 1988]
XX
AN 176
NM GRE(1); glucocorticoid responsive element
NS R00591
TF GR; glucocorticoid receptor [Akerblom et al., 1988]
AT decrease
SQ AGATCAAATTGACGT
PQ -151 to -137 (C)
BF HSGPHA:185
AG 1.1.5, 4.2, 6.1.1+, 7.1 [Akerblom et al., 1988]
XX
AN 173
NM CRE(1);cAMP response element
NS R00212, R00592
TF CREB; cAMP response element binding protein [Bokar J.A. et al.,1989,
TF Deutsch P.J. et al., 1988]
AT increase
SQ TGACGTCA
PQ -142 to -135
BF HSGPHA:
AG Placenta, pituitary cells: 3.4, 6.1, 6.3 [Bokar J.A. et al., 1989]
AG 3.2.1, 3.3, 6.2+, 6.4+, 6.5+, 7.3 [Deutsch P.J. et al., 1988]
AG Placenta, pituitary cells: 1.1.1, 1.1.2, 3.4, 3.6, 6.2
AG [Steger et al., 1994]
HM Bovine a-GH, CRE, -139 to -132, TGATGTCA,
HM Mouse a-GH, CRE, -139 to -132, TGATGTCA,
HM Rat a-GH, CRE, -136 to -129, TGATGTCA. [Bokar J.A. et al., 1989]
HM CREs of rat glucagon, bovine PTH, human enkephalin TG-CGTCA, human
HM VIP, rat somatostatin [Deutsch P.J. et al., 1988]
EF The affinity of the bovine a-GH CRE element for binding with CREB
EF is at least 200 times lower than that of the human a-GH CRE
EF [Bokar J.A. et al., 1989]
EF The affinity of the a-GH CRE for binding with CREB is
EF indistinguishable from the affinity of the rat som.CRE and much more
EF than that of the bov.PTH, rat glucagon and mutant a-GH CRE,
EF containing central C to G transversion [Deutsch P.J. et al., 1988]
XX
AN 174
NM CRE(2); cAMP response element
NS R00212, R00592
TF CREB; [Bokar J.A. et al., 1989, Deutsch P.J. et al., 1988]
AT increase
SQ TGACGTCA
PQ -124 to -117
BF HSGPHA:
AG Placenta, pituitary cells: 3.4, 6.1, 6.3 [Bokar J.A. et al., 1989]
AG 3.2.1, 3.3, 6.2+, 6.4+, 6.5+, 7.3 [Deutsch P.J. et al., 1988]
KK consensus sequence of CRE is a palindrome [Deutsch P.J. et al., 1988]
XX
AN 175
NM JRE; junction regulatory element
NS R02847
TF JRF; junction regulatory factor [Andersen et al., 1990]
AT increase
SQ GTAATTAC
PQ -114 to -107
BF HSGLYHAS:731
BF HSGPHA:216
BF HSGPHAA:196
AG 1.1.1, 1.1.2, 3.4, 4.2, 6.2 [Andersen et al., 1990]
HM JRE is conserved between human, rat, mouse, cattle
HM [Andersen et al., 1990]
KK sequence of the JRE is a palindrome [Andersen et al., 1990]
CC JSO is placenta-specific 50 kD factor [Andersen et al., 1990]
XX
AN 177
NM StRE; steroid-responsive element
NS R00214, R00593
TF AR; androgen receptor [Clay C.M. et al., 1993] +
TF GR; glucocorticoid receptor [Akerblom et al., 1988]
AT decrease
SQ ATTACACCAAGTACCCTTCAAT
PQ -111 to -90 (C)
BF HSGPHA:234
AG 3.2.1, 3.2.2, 3.3, 3.4, 3.5, 7.1 [Clay C.M. et al., 1993]
AG 1.1.5, 4.2, 6.1.1+, 7.1 [Akerblom et al., 1988]
XX
PR proximal promoter; ST; -90 to +1; 179, 180, 181
PR [Bokar et al., 1989, Chatterjee et al., 1989]
XX
AN 179
NM CCAAT box; (C)
NS R02848
TF a-CBF; alpha subunit CCAAT-binding factor [Kennedy et al., 1990]
AT increase
SQ CCAAT
PQ -87 to -83
AG 3.3, 3.4, 3.5, 4.2, 6.2+ [Kennedy et al., 1990]
EF a-CBF binds to the human alpha-gh CCAAT box with at least a 100-fold
EF greater affinity than to other CCAAT boxes [Kennedy et al., 1990]
KK Human a-CBF has an apparent molecular mass of 53 kD and distinct from
KK CTF/NF1, C/EBP, CP1, and NF-Y. alpha-CBF is present in a variety of cell
KK types. It is likely that the binding of a-CBF to the CCAAT element
KK serves as a bridge between placenta-specific enhancer and TATA-binding
KK proteins [Kennedy et al., 1990]
XX
AN 178
NM GRE(2); glucocorticoid responsive element
NS R00595
TF GR; glucocorticoid receptor [Akerblom et al., 1988]
AT decrease
SQ ATTTCCTGTTGATCC
PQ -79 to -65
BF HSGPHA:258
AG 1.1.5, 4.2, 6.1.1+, 7.1 [Akerblom et al., 1988]
XX
AN 180
NM TATA box;
SQ TATAAAA
PQ -29 to -23
AG 7.1 [Bokar J.A. et al., 1989]
XX
AN 181
NM TRE; thyroid responsive element
TF erbA-beta TR; erbA-beta form of thyroid hormone receptor
TF [Chatterjee et al., 1989]
AT decrease
SQ GCAGGTGAGGACTTCA
PQ -22 to -7
AG 3.2.1, 3.5, 6.1.1+, 6.1.2+, 7.2 [Chatterjee et al., 1989]
HM TRE sites of rGH [Chatterjee et al., 1989]
XX
AL Bovine a-GH, sequence identity of 85%, -313 to -1 [Bokar et al., 1989]
XX
CC Each part of 18 bp direct repeat (-146 to -129 and -128 to -111)
CC contains CRE consensus. [Bokar et al., 1989]
XX
CC The mutation at the 5'-boundary of CRE, T --> A, deminished both
CC basal and cAMP stimulated activity by 95 %,
CC activity in the CRE 3' A to T mutant was below the level of
CC detection. [Deutsch P.J. et al., 1988]
XX
CC Placenta-specific expression of alpha-subunit gene is provided by
CC complex interaction of at least four different elements (URE,
CC CRE, JRE, and CCAAT) with each binding a distinct
CC protein. [Andersen et al., 1990]
XX
AU Deutsch P.J., Hoeffler J.P., Jameson J.L., Lin J.C., Habener J.F.
TI Structural determinants for transcriptional activation by cAMP
TI responsive DNA elements.
SO J. Biol. Chem. (1988), v. 263, n. 34, 18466-18472
//
1. Miltenberger R.J., Sukow K.A., and Farnham P.J. // Mol. Cell. Biol. 1995. V. 15. P. 2527-2535.
2. Karlseder J., Rotheneder H., and Wintersberger E. // Mol.Cell.Biol. 1996. V. 16. P. 1659-1667.
3. Zhu L., Zhu L., Xie E., and Chang L.-S. // Mol. Cell. Biol. 1995. V. 15. P. 3552-3562.
4. Tsai E. Y., Jain J., Pesavento P.A., Rao A., and Goldfeld A.E. // Mol. Cell. Biol. 1996. V.16. P459-467.
5. Jackson D.A., Rowader K.E., Stevens K.Y., Jiang C., Milos P., and Zaret K.S. // Mol.Cell.Biol. 1993. V. 13. P. 2401-2410
6. Li W.-H., Tanimura M., Luo C.-C., Datta S., Chan L.//Journal of Lipid Research. 1988. V. 29. P. 245-271
7. Knott T.J., Rall S.C., Innerarity T.L., Jacobson S.F., Urdea M.S., Levy-Wilson B., Powell L.M., Pease R.J., Eddy R., Nakai H., Byers M., Priestley L.M., Robertson E., Rall L.B., Betsholtz C., Shows T.B., Mahley R.W., Scott J. // Science. 1985. V. 230. P. 37-43.
8. Cladaras C., Hadzopoulou-Cladaras M., Avila R., Nussbaum A., Nikolosi R., Zannis V.I. // Biochemistry. 1986. V. 25. P. 5351-5357.
9. Zannis V.I., Cole F.S., Jacson C.L., Kurnit D.M., Karathanasis S.K. // Biochemistry. 1985. V. 24. P. 4450-4455
10. Knezetic J.A. and Felsenfeld G. // Mol.Cell.Biol. 1993. V. 13,.P. 4632-4639
11. Meier V.S. and Groner B. // Mol. Cell. Biol. 1994. V. 14. P. 128-137.
12. Steger D.J., Hecht J.H., and Mellon P.L. // Mol.Cell.Biol. 1994. V. 14. P5592-5602
13. McMahon S.B. and J.G. Monroe. // Mol. Cell. Biol. 1995. V. 15. P. 1086-1093.
14. Kel O.V., Romachenko A.G., Kel A.E., Naumochkin A.N., Kolchanov N.A. Proceedings of the 28th Annual Hawaii International Conference on System Scienses [HICSS]. 1995. V. 5. Biotechnology Computing, IEE Computer Society Press, Los Alamos, California,P. 42 -51
15. Kolchanov N.A., Mol. Biol., 1997, vol. 31, pp. 581-583.
16. Roeder R.G. // Trends Biochem. Sci. 1996. V. 249. P. 327-335.
17. Bjoerklund S., Kim Y.-J. // Trends. Biochem. Sci. 1996. V. 249. P. 335-337.
18. Kaiser K. and Meisterrernst M.// Trends Biochem. Sci. 1996. V. 249. P. 342-345.
19. Verrijzer C.P., Tjian R. // Trends Biochem. Sci. 1996.V. 249. 338-342.
20. Wingender E. Gene regulation in eukaryotes. Germany:VCH, 1993. 430p.
21. Kel O.V., A.G.Romaschenko, A.E.Kel, E.Wingender, N.A.Kolchanov. A //. Nucl. Acids Res. 1995. V.23. P. 4097-4103.
22. Ferreri K., Gill G., and Montminy M. // Proc. Natl. Acad. Sci. USA. V. 91. P. 1210-1213.
23. Metz R., Bannister A.J., Sutherland J.A., Hagemeier C., O'Rourke E.C., Cook A., Bravo R., and Kouzarides T. // Mol. Cell. Biol. 1994. V. 14. P. 6021-6029.
24. MacDonald P.N., Sherman D.R., Dowd D.R., Jefcoat S.C., and DeLisle R.K. // J. Biol. Chem. 1995. V. 270. P. 4748-4752.
25. Xing L., Venkatesh K.G., and Quinn P.G.. // J. Biol. Chem. 1995. V. 270. P. 17488-17493.
26. Schmitz M.L., Stelzer G., Altmann H., Meisterernst M., and Baeuerle P.A. // J. Biol. Chem. 1995. V. 270. P. 7219-7226.
27. Hernandez-Munain C. and Krangel M.S. // Mol. Cell. Biol. 1995. V. 15. P. 3090-3099.
28. Kamachi Y. and Kondoh H..// Mol.Cell.Biol. 1993. V. 13. P. 5206-5215
29. Pongubala J.M.R., Nagulapalli S., Klemsz M.J., McKercher S.R., Maki R.A., Atchison M.L. // Mol. Cell. .Biol. 1992. V. 12. P. 368-378
30. Dynan W.S. // Cell, 1989, V 58, P1-4,
31. Wingender E., Dietze P., Karas H., and Knuppel R. // Nucl. Acids Res. 1996. V. 24. P. 238-241.
32. Anan'ko E.A., Bazhan S.I., Belova O.E., and Kel' A.E., Mol. Biol., 1997, vol. 31, pp. 701-713.
33. Ignat'eva E.V., Merkulova T.I., Vishnevskii O.V., and Kel' A.E., Mol. Biol., 1997, vol. 31, pp. 684-700.
34. Merkulova T.I., Merkulov V.M., and Mitina R.L., Mol. Biol., 1997, vol. 31, pp. 714-725.
35. Podkolodnaya O.A. and Stepanenko I.L., Mol. Biol., 1997, vol. 31, pp. 671-683.
36. Kel' O.V., Kel' A.E., Romashchenko A.G., Wingender E., and Kolchanov, N.A., Mol. Biol., 1997, vol. 31, pp. 601-615.
37. Wade R., Kedes L. // Ann. Rev. Physiol. 1989. V. 51. P. 179-188.
38. Rhodes S.J., DiMattia G.E., Rosenfeld M.G. // Carrent Opin. In Genetics and Development. 1994. V.4. P. 709-717.
39. Ray A., Hannink M., and Ray B.K. // J. Biol. Chem. 1995. V. 270.P. 7365-7374.
40. P.Bucher, EPD (Eukaryotic Promoter Database) current release
41. Kolpakov F.A. and Babenko V.N., Mol. Biol., 1997, vol. 31, pp. 647-655.
42. Berg P.E., Willams D.M., Qian R.L., Cohen R.B. Cao S.-X.,Mittelman M., Schachter A.N. // Nucl. Acids Res. 1989, V 17, P 8833 - 8852.
43. Wall L., deBoer E., Grosveld F. // Genes and Dev. 1988, V 2, PG1089 - 1100.