THE FUNCTIONAL SITES OF PRO- AND EUKARYOTIC GENOMES:
MODELS FOR ACTIVITY PREDICTION
Dear Chairman, dear colleagues!
I would like to offer you a new computer technology (Fig. 1) for prediction of functional site activity on the basis of their nucleotide sequences. As is known, every molecular process in a cell, such as replication, transcription, splicing, and translation, is controlled by a definite set of functional sites. Functional sites of the same type can differ in their activity by several orders of magnitude. For example (Fig. 2), the 3'end pre-mRNA processing efficiency of SV40 virus varies in the range of one order of magnitude; E. coli binding sites differ 100-fold in the affinity for Cro repressor; E. coli binding sites differ 350-fold in the affinity for RecA protein, and so on.
A number of approaches have been proposed to predict the activities of the sites on the basis of their contextual characteristics (Fig. 3), namely, consensus, weighted matrix, multiple regression, neural networks, and others. All the approaches employ "SEQUENCE - ACTIVITY" table of experimental data as initial information for the analysis (Fig. 4). Note, however, that currently there are hundreds of such site samples with experimentally determined activity levels. Nevertheless, the sequence-based predictions of site activity have rarely been successful. Therefore, the prediction of the site activity remains a challenging problem of computational biology.
Our approach (Fig. 5) is new in molecular-biological terms because in addition to the traditional contextual characteristics it takes into account physical-chemical and conformational DNA properties. Our approach is new also in the mathematical terms because it uses methods for generation and testing of hypothesis within the framework of the Utility Theory for Decision Making together with Zadeh's fuzzy logics.
Before discribing our approach, let me show you three examples of its capacities. The performed analysis demonstrated (Fig. 6) that in prediction of the efficiency of the 3'-end pre-mRNA processing, it is most important to take into account the weighted concentration of the tetranucleotide VUKK located to the right of the RNA cleavage site. In prediction of the USF-protein affinity for DNA (Fig. 7), another property is important, namely such DNA conformational characteristics as the twist angle. As for prediction of the CRO-repressor affinity for DNA, such conformational characteristic of DNA as the major groove width was shown to be the most significant
(Fig. 8).
The scheme of the ACTIVITY computer system is shown in this slide (Fig. 9). An important unit of the system is the database on site activity. The current version of the database contains more than 70 sample sites of different types with experimentally determened activity levels (Fig. 10). Among them are promoters and binding sites for different E. coli regulatory proteins, TATA-boxes and binding sites for various eukaryotic transcription factors, translation initiation sites, and others. The parameters characterizing specific site activities include the association and dissociation rate constants of DNA-protein complexes, equilibrium constants and the half-life periods of the complexes, and various quantitative values characterizing the yield of the end products controlled by these sites, for example, their promoter strength and translation efficiency.
The core idea of our approach (Fig. 11) implies that the site activity F is determined by simultaneous action of characteristics of two types: obligatory and facultative. Obligatory characteristics are the same for all sequence variants of a given site; they determine the basal activity level of the sites of a given type. The facultative characteristics are individual for each sequence variant of a given site (in terms of their number and location). They modulate individual site activity with respect to basal level. Then, within the framework of the linear-additive model, the activity of the site Sn is described by the following equation (Fig. 12):
![]() ;
;
Here, F0 is the basal activity level of the sites of a given type; Fk is the contribution of the facultative characteristic Xk to the activity F. Xk(Sn) is the value of the kth facultative characteristic of the sequence Sn.
Three types of facultative characteristics are considered in the Activity system: statistical, conformational, and physical-chemical. The weighted concentrations of oligonucleotides are considered as statistical characteristics of the nucleotide context of the sites. They are calculated (Fig. 13) as follows:
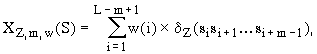
Here, d is the so-called indicator function:
![]()
The function ![]() describes the distribution of the
oligonucleotide Z along the site sequence. At a definite position, it assumes the value
"1" or "0" depending on the presence or absence of oligonucleotide Z
at this position.
describes the distribution of the
oligonucleotide Z along the site sequence. At a definite position, it assumes the value
"1" or "0" depending on the presence or absence of oligonucleotide Z
at this position.
The basic element of the facultative characteristic description (Fig. 14) is the function of position effect W(i). The function allows to take into consideration the fact that the same oligonucleotide contributes differently to the site activity depending on its location The function w(i) is determined by a simple rule: the more important is the position for the site function, the higher is its assigned weight w(i). The total number of the weighted functions W(i) used in the activity prediction is 200. The weight functions given here (Fig. 14) demonstrate the highest effect on the site activity of (a) the right half of the sequence, (b) its central part, (c) its terminal regions, and (d) the narrow region within the right half of the site.
It should be noted that oligonucleotides are considered in the expanded 15 single-letter base codes, which have an unambiguous physical-chemical sense (Fig. 15).
We also use DNA conformational properties in the prediction of site activity. The conformational properties play an important role in DNA-protein interactions, which essentially determine the site activity (Fig. 16). Local conformational heterogeneities of DNA and its dependence on the nucleotide context are the characteristic features of double-stranded DNA. The local conformational parameters determine the mutual orientation of base pairs and other conformational features of DNA. X-ray analysis of DNA and DNA-protein complexes allowed us to assign the average conformational parameters for each dinucleotide. They are stored in a special database of the Activity system (Fig. 17a). For example, propeller twist is a rotation angle between two bases along the long axis of the complementary pair. The dinucleotides GT and AC have the smallest propeller twist, whereas the AA dinucleotide has the largest value (Fig. 17a). About 40 conformational properties of DNA are used in prediction of the site activities (Fig. 18).
Similarly, the mean values of a number of physical-chemical properties are also assigned to each dinucleotide, such as the DNA melting temperature (Fig. 17b), persistent DNA length, DNA flexibility, entropy, and others. These properties determine the conformational dynamics of DNA sites during their functioning and for this reason they are used in prediction of the site activities (Fig. 19)
Thus (Fig. 20), the sequence of the site S can be characterized by the mean value of the qth conformational or physical-chemical propertiy of DNA in the region (a, b):
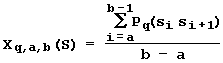 .
.
It should be emphasized that BEFORE starting the analysis, we knew very little about the statistical, physical-chemical, or conformational features that were most important for the activity of the examined site. In most cases, even approximate information about the regions of a site that are most important for its activity is lacking. The only available data are the tables listing the sequences with the known activities (Fig. 21). With this in mind, the basic principle of the Activity system is the principle of impartiality. This principle is fundamental in the artificial intellegence systems (Fig. 22). The idea is the following. When the information is insufficient, the more hypotheses have been tested, the more correct is the result. No preference is given to any hypothesis before its testing. In our case, the hypothesis is the assumption that each particular statistical, conformational, or physical-chemical characteristic calculated by the above approach is significant for the activity of the examined site.
For this reason, in the analysis of statistical characteristics, we tested, one by one, all the variants of oligonucleotide Z varying (a) its length from 1 to M; (b) its nucleotide composition in 15 single-letter based codes; and (c) all available functions of position effect w(i) (Fig. 23). The weighted concentration of oligonucleotide Z is calculated for fixed combinations <Z,m,w> for each sequence S. Here, the total number of combinations is about 107. Similarly (Fig. 24), for each fixed conformational or physical-chemical parameter, all the possible regions (a, b) within the site are considered, one by one. Then, for a fixed combination of parameters and region, the mean value of the parameter is calculted for each sequence S. The total number of combinations here is about 105.
The validity of each hypothesis in determination of the contribution of each considered characteristic to the site activity is estimated (Fig. 25) on the basis of approaches developed within the framework of the Fishburn's Theory of Utility for Decision Making. In turn, this theory is based on the concept of fuzzy calculations developed by Zadeh. In our case, the U value, the utility of a characteristic in prediction of the site activity, is calculated for each examined characteristic Xzm(Sn).
Let us calculate the characteristic Xzmw(Sn) for each sequence Sn with the known activity Fn. If the pairs {Xzmw(Sn), Fn} meet the conditions of regression analysis, then Fn can be predicted from Xzmw. To check this, a simple regression is used:
![]() ;
;
To ensure the reliability of the regression between the Xzmw(Sn) and Fn values (Fig 25a.), 22 conditions of regression analysis are tested, namely, the presence of linear, sign, and rank correlations between the predicted and experimentally observed activities; the equality of distributions of these values, and so on. When testing each of the 22 conditions, the significance level at which a condition is met is estimated.
At the next step (Fig. 26), the partial utility of characteristic XZmw for the prediction of the activity F is calculated as follows:
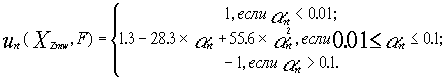
The highest partial utility U=1 is assigned to the characteristic Õ
zmw, if the corresponding criterion is met at significance level a rt.less then 0.01. The utility is lowest (that is -1), if the corresponding criterion is not met (a rt exceeding 0.1). At the intermediate a rt value, the corresponding intermediate U value from the interval [-1 to +1] is assigned to the characteristic Õzmw. On the basis of the 22 partial utilities, the integral utility of the characteristic Õzmw for the prediction of the activity F is calculated as: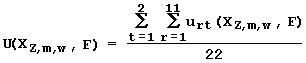 .
.
To predict the site activity, only characteristics with positive integral utility are selected (U(XZmw,F) > 0) (Fig. 27). Among them, a limited set of the linearly independent characteristics with the highest U value is selected. Each such selected characteristic possesses two helpful properties: (1) it correlates with the activity level and (2) it does not correlate with any other selected characteristic. The probability that the characteristic was randomly selected, estimated by the binomial distribution (Fig. 28), is less than 10-9.
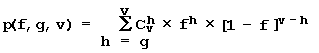
Shown in the next slide (Fig. 29) are the results of analysis of E. coli promoters with known activity levels. The weighted concentration of the trinucleotide ASM was shown to be the most significant for the promoter strength. The corresponding weight function w(i) assigns the highest significance to the site locations within -1 to +11 region of the promoter. This means that this particular region is the most important for the contribution of the trinucleotide ASM to the promoter strength. There is a significant correlation between the weighted ASM concentration and promoter strength (Fig 29a). One more important characteristic (Fig. 30) of promoter strength is the mean value of the direction angle in the region [-4 to +16] of the promoter. There is a significant correlation between this characteristic and the promoter strength. Basing on these characteristics, the canonical equation (1) for predicting site activity is derived (Fig. 31). This equation provides a significant agreement between the experimental data and calcualted values of promoter strength. The text (Fig. 32) of the program allowing to calculate the activity F(S) of the site with the arbitrary sequence S is automatically generated by the Activity system. It is recorded in the special program library of the Activity system available through the Internet.
Some dozens of natural sites of different types were analyzed by the Activity system, and the results demonstrated the universality of this approach. In all the cases, the significant statistical, physical-chemical, or conformational characteristics were identified and the methods for sequence-based prediction of site activity were developed.
Analysis of the sequences with known DNA bending in the ÒÂÐ/ÒÀÒÀ complex demonstrated that the bending increases (Fig. 33) with the inclination angle of free DNA. Similar results were obtained by the X-ray analysis of the TBP/TATA complexes. DNA bending in these complexes was shown to result from intercalation of four phenylalanine residues of the ÒÂÐ between a pair of adjacent bases at the side of the minor groove (Fig. 34). Inclination describes the rotation angle of a pair of bases along the short axis of the pair, and the increase in the angle widens the minor groove, thereby facilitating the intercalation of phenylalanines at the side of the minor groove and consequent DNA bending.
This approach can be applied also to synthetic analogues of sites and their mutational variants. Study of the synthetic analogues of the ÒÀÒÀ-boxes with known TBP affinity revealed two statistically significant characteristics (Fig. 35): (1) the weighted concentration of the dinucleotide TV, contributing primarily to the TBP affinity in the centre of the site; and (2) the weighted concentration of the dinucleotide WR, chiefly contributing to the affinity at the site termini. The prediction method for TBP affinity developed on the basis of these characteristics demonstrates a good agreement between the predicted and experimentally observed values for the DNA/TBP affinity.
Analysis of mutational variants of mouse aÀ-crystalline gene promoter (Fig. 36) has shown that the most important characteristic of this region is the frequency of the contacts of dinucleotides of the promoter with nucleosome core proteins. This characteristic negatively correlated with transcriptional activity. This means that the tighter is the interaction of the promoter with nucleosomes, the lower is the transcription level of aÀ-cristallin gene. This result is consistent with the experimental data showing that nucleosome displacement from the promoter region precedes the ÒÂÐ/ÒÀÒÀ-binding. The analysis performed demonstrated also (Fig. 37) that such conformational characteristics of DNA as the TILT angle and the parameter DIST are of great importance for the transcription efficiency of the promoter. Using these three characteristics (Fig. 38), a method was developed to predict the transcriptional activity of the aÀ-crystalline gene promoters on the basis of their sequences.
Summing up, I would like to underline that the Activity system is applicable to a wide range of experimental data on site activity however requiring their minimal volume and is completely automated. The computer Activity system is available through the Internet (Fig. 39). Thus, the Activity system has a set of tools to be successfully used for studying functional sites.