MATHEMATICAL MODEL OF CHOLESTEROL BIOSYNTHESIS REGULATION IN THE CELL
*Ratushny A.V., Ignatieva E.V., Matushkin Yu.G., Likhoshvai V.A.
Institute of Cytology and Genetics SB RAS, Russia
e-mail:ratushny@bionet.nsc.ru
*
Corresponding author,
Keywords: gene network, cholesterol, regulation, mathematical model, computer analysis
Resume
Motivation:
An adequate mathematical model of the complex nonlinear gene network regulating cholesterol synthesis in the cell is necessary for investigating its possible function modes and determining optimal strategies of its correction, therapeutic included.
Results:
Dynamic model of function of the gene network regulating cholesterol synthesis in the cell is constructed. The model is described in terms of elementary processesóbiochemical reactions. The optimal set of parameters of the model is determined. Patterns of the system behavior under different conditions are simulated numerically.
Introduction
Cholesterol, an amphipathic lipid, is an essential structural component of cell membranes and outer lipoprotein layer of blood serum. In addition, cholesterol is a precursor of several other steroids, namely, corticosteroids, sex hormones, bile acids, and vitamin D. Cholesterol is synthesized in many tissues from acetyl-CoA and its main fraction in blood serum resides with low-density lipoproteins (LDL). Free cholesterol is removed from the tissues with involvement of high-density lipoproteins (HDL) and transported to the liver to be transformed into bile acids. Its major pathological role is in serving as a factor causing atherosclerosis of vital cerebral arteries, heart muscle, and other organs. Typical of coronary atherosclerosis is a high ratio of LDL to HDL cholesterol [Marry R. et al., 1993]. Haploid and diploid versions of the dynamic model of function of the gene network regulating cholesterol synthesis in the cell are constructed in the work. The models are described in terms of elementary processesóbiochemical reactions. The optimal set of parameters of the model allowing the calculations to comply with the published experimental data is determined through numerical experiments. Patterns of the system dynamic behavior under different conditions are simulated numerically. The results obtained are compared with the available experimental data.
Cholesterol biosynthesis and its regulation
Approximately half of the cholesterol amount present in the organism is formed through biosynthesis (about 500 mg/day) [Marry R. et al., 1993], while the other half is consumed with food. The main part of cholesterol is synthesized in the liver (~ 80% of the total cholesterol produced), intestines (~ 10%), and skin (~ 5%) [Klimov & Nikulícheva, 1999].
Acetyl-CoA is the source of all the carbon atoms composing the cholesterol molecule. The main stages of cholesterol biosynthesis are described in the GeneNet database.
Cholesterol regulates its own synthesis and the synthesis of LDL receptors at the level of transcription through a negative feedback mechanism [Wang et al., 1994]. A decrease in the cell cholesterol content stimulates SRP (sterol regulated protease)- catalyzed proteolysis of the N-terminal fragment of SREBP (sterol regulatory element- binding protein), bound to the endoplasmic reticulum (ER) membrane. On leaving the ER membrane, SREBP migrates to the cell nucleus to bind the so-called sterol regulatory element (SRE), residing in the promoter of the receptor gene, thereby switching on the receptor synthesis. In addition, SREBP activates the gene of synthase of hydroxymethyl glutaryl (HMG)-CoA reductase [Klimov & Nikulícheva, 1999] as well as farnesyl diphosphate synthase and squalene synthase syntheses. Several studies have demonstrated rather fast effect of cholesterol on the reductase activity, unexplainable by the mere effect on the rate of enzyme synthesis. HMG-CoA reductase may be either active or inactive. Phosphorylation- dephosphorylation reactions provide for the transitions from one state into the other [Marry R. et al., 1993].
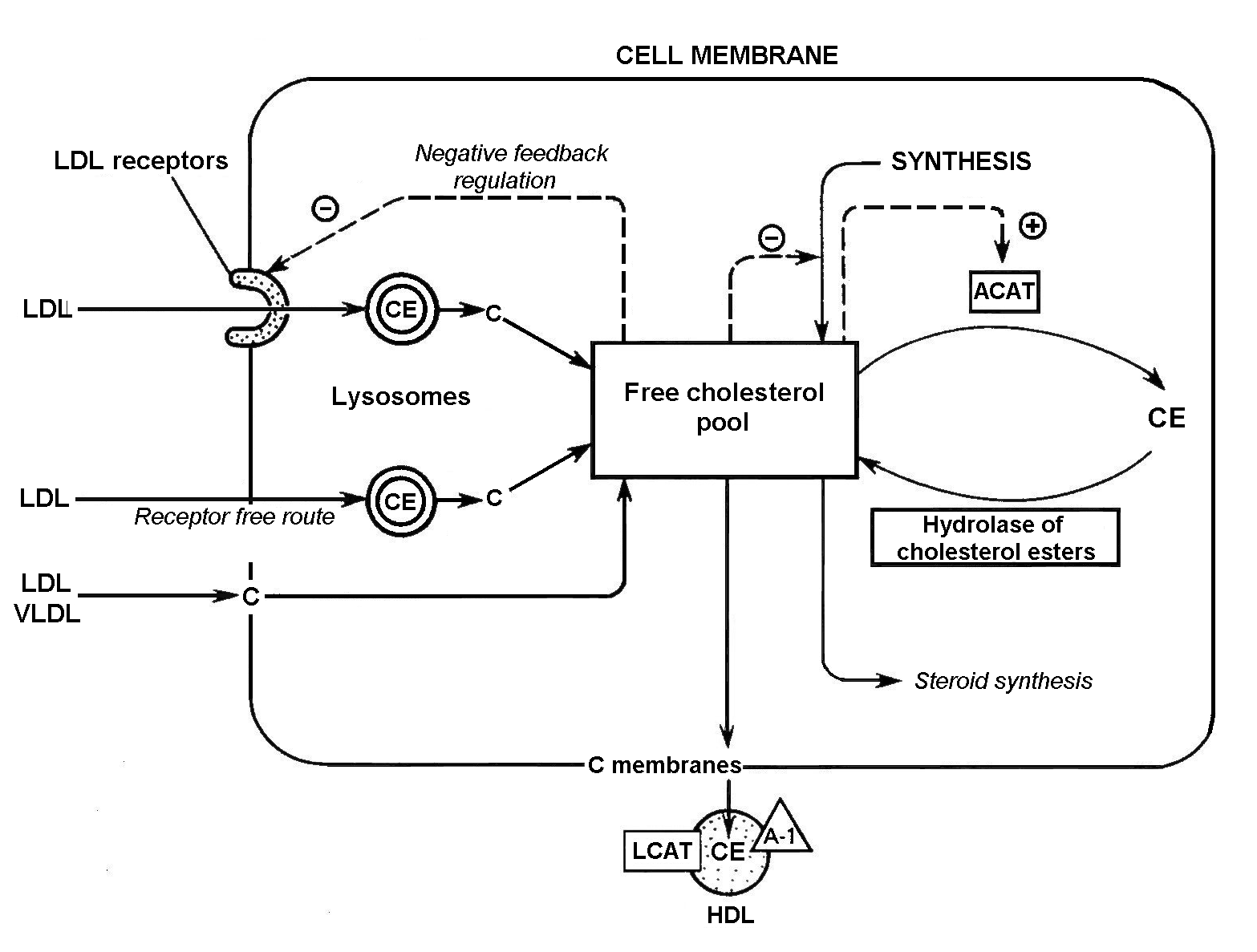
The main factors affecting the cholesterol balance at the cell level [Marry R. et al., 1993] are shown in Fig. 1.
Figure 1. Factors affecting the cholesterol balance at the cell level: C, cholesterol; CE, cholesterol esters; ACAT, acyl-CoA:cholesterol acyltransferase; LCAT, lecithin:cholesterol acyltransferase; A1, apoprotein A1; LDL, low density lipoproteins; VLDL, very low density lipoproteins, HDL, high density lipoproteins; (- ), inhibition of cholesterol synthesis; and (+) ACAT activation [Marry R. et al., 1993].
Cell cholesterol content increases if (1) specific LDL receptors bind cholesterol-containing lipoproteins; (2) cholesterol-containing lipoproteins are bound without receptors; (3) free cholesterol, contained in cholesterol-rich lipoproteins is bound by cell membranes; (4) cholesterol is synthesized; and (5) cholesterol ester hydrolase- catalyzed hydrolysis of cholesterol esters takes place.
Cell cholesterol content decreases if (1) cholesterol passes from membranes into cholesterol-poor lipoproteins, in particular LDL3 or LDL synthesized de novo (lecithin:cholesterol acyltransferase promotes this transition); (2) ACAT-catalyzed cholesterol esterification takes place; and (3) cholesterol is used for synthesizing other steroids, in particular, hormones or bile acids in the liver [Marry R. et al., 1993].
Methods and algorithms
A generalized chemical kinetic approach [Bazhan et al., 1995] was used for the simulation. A blockwise formalization was used, that is, each process is separated in an individual block and described independently of the other processes. A block is a simulation quantum, and its formal structure is completely described with the following three vector components: (1) X, the list of dynamic variables; (2) P, the list of constants; and (3) F, type of the right part of the system dX/dt = F(X, P) determining the rule these dynamic variables change with time. Four types of blocks are used to describe the processes in the model, namely:
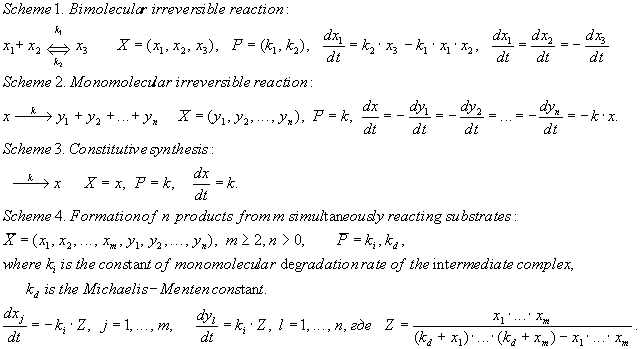
Successive application of the blockwise approach to description of biological systems is based on the law of summation of the rates of elementary processes while uniting them in a general scheme of the simulated object development. The method of Gear [Gear, 1971] was used for numerical integration of the set of differential equations.
Results
Mathematical model
The mathematical model of intracellular cholesterol biosynthesis regulation comprises 65 kinetic blocks, 40 dynamic variables, and 93 reaction constants. The diploid model comprises 72 kinetic blocks, 44 dynamic variables, and 130 reaction constants. Experimental data, partially listed in table below, were used for the initial evaluation of certain parameters of enzymatic reactions with the system.
Table. Some constants of enzyme reactions
Enzyme |
Substrate |
Organism |
Organ |
Kc, sec- 1 | Km, mM |
| HMG-CoA reductase | HMG-CoA | Rattus norvegicus [Gil et al., 1981] | Liver | 980 |
(-) |
| HMG-CoA reductase | HMG-CoA | Rattus norvegicus [Kleinsek & Porter, 1979] | Liver | (-) |
0.0169 |
| HMG-CoA reductase | HMG-CoA | Rattus norvegicus [Sugano et al., 1978] | Intestine | (-) |
0.0417 |
| HMG-CoA synthase | Acetyl-CoA Acetoacetyl-CoA |
Gallus gallus (hen) [Reed et al., 1975] | Liver | (-) (-) |
0.1? 0.7 <0.005 |
| HMG-CoA synthase | Acetyl-CoA | Homo sapiens [Rokosz et al., 1994] | Adrenal | (-) |
0.029 |
| Acetoacetyl-CoA thiolase | Acetoacetyl-CoA CoA |
Bos taurus (calf) [Huth et al., 1975] | Liver | (-) (-) |
0.01 0.025 |
| Acetoacetyl-CoA thiolase | Acetoacetyl-CoA CoA |
Gram-negative bacteria [Kim & Copeland 1997] | 2.38e+4 2.38e+4 |
0.042 0.056 |
|
| Presqualene synthase | Farnesyl diphosphate | Saccharomyces cerevisiae (yeast) [Sasiak & Rilling, 1988] | (-) |
0.03 |
|
| Geranyltransferase | Geranyl PP Isopentyl PP |
Homo sapiens [Barnard & Popjak 1981] | Liver | 40.7 40.7 |
4.4e-4 9.4e-4 |
| Lanosterol synthase | (R,S)-squalene-2,3-oxide | Saccharomyces cerevisiae [Balliano et al., 1992] | (-) |
0.035 |
|
| ACAT-1 | Oleoyl-CoA Cholesterol |
Homo sapiens (Cricetulus griseus)[Chang et al., 1998] | Ovary | (-) |
7.4Ś-3 |
| Bile acid hydrolase | Taurocholate | Lactobacillus sp. (bacteria) [Lundeen & Savage, 1990] | 1900 |
0.76 |
Other published data were used for evaluating parameters of the model, in particular [Klimov & Nikulícheva, 1999]:
Fasting LDL concentration in adult human blood serum CLDL = 200- 300 mg/dl.
The average number of unesterified and esterified cholesterol molecules per one LDL particle QUEC = 475 and QEC = 1310.
LDL half-life in blood of healthy humans t 1/2 = 2.5 days; therefore, kLDLutil. = ln(2)/t 1/2 =3.21*10-6 sec-1.
Total number of LDL receptors per one cell at 37į C QLDLR = 15,000- 70,000.
Lifespan of LDL receptors t = 1- 2 days; therefore, kLDLRutil. = 1/t ~ 7.72e - 6 sec- 1.
LDL receptor recyclization span t ~ 20 min.
The values of the rest parameters of the model were determined through numerical experiments.
Figure 2. Kinetics of main components of the system regulating cholesterol biosynthesis in the cell.
Results of calculations
The results obtained while simulating the cell response to a twofold increase in LDL particle content in blood serum (Fig. 2, b) illustrate the model performance. The number of receptors bound to LDL increases (d); unbound, decreases (e). Intracellular concentrations of free cholesterol (a) and its esters (c) increase. Free cholesterol binds the protease (SRP), preventing SREBP-1 formation (f). Productions of enzymes involved in the internal cellular cholesterol synthesis (HMG-CoA reductase; g), LDL receptors, and intermediate low-molecular-weight components (mevalonic acid, h; squalene, i) are stopped. Cholesterol concentration in the cell is decreasing. No further influence on the system provided, it returns to the initial state. A complete recovering requires about 15 h.
In future, we plan to perform computer stimulation of recombination process in diploid cell, by modelling interactions between alleles of genes responsible for cholesterol biosynthesis.
Acknowledgments
The authors are grateful to Galina Chirikova for translation of the manuscript into English and to N.A. Kolchanov for fruitful discussions. The work was supported by National Russian Program "Human Genome" (No 106), Integrational Science Project of SB RAS "Modelling of basic genetical processes and systems".
References