Activity
General description
The identification of the sequence-dependent DNA features correlating with
affinity magnitudes of DNA sites interacting with a protein can pinpoint to
molecular event limiting this protein/DNA recognition machinery. The approach is
realized in computer system ACTIVITY containing the databases on site activity
and on conformational and physical-chemical DNA/RNA parameters. By using the
system ACTIVITY, an analysis of some sites was provided and the methods for
predicting the site activity were constructed.
How to Run the Program
1. Input the DNA sequence of interest into the field
"Input DNA Sequence". For example, recognition of the promoter of
Syrian hamster HMG CoA reductase gene (EMBL AC M15960), the experimental USF
site TRANSFAC: R00711 (181; 197) is illustrated.
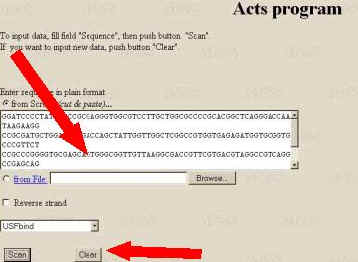
2. Select one of transcription factor binding sites from
the list and denote the strand.
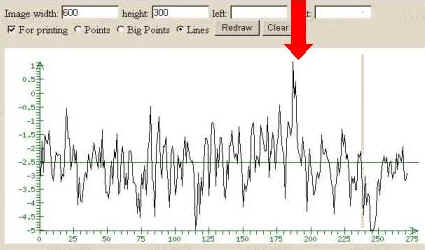
3. Start the tools processing by clicking the button
"Scan". The tools output represents the profile of the Score value.
The positive peaks of this profile could point to the potential site
recognized.
Ponomarenko J.V., Furman D.P., Frolov A.S., Podkolodny N.L., Orlova G.V.,
Ponomarenko M.P., Kolchanov N.A., and Sarai A. (2001) ACTIVITY: a database on
DNA/RNA sites activity adapted to apply sequence-activity relationships from one
system to another Nucleic Acids Res.,V.29. N.1. P. 284-287.

