Freq Matrix
General description
FreqMATRIX is a Web-available resource describing the site-specific oligonucleotide content of the functional DNA sites and applying it for the site recognition. To describe specific oligonucleotide content of the functional DNA sites, the oligonucleotide alphabets were introduced. Out of these alphabets, the frequency matrix for a given site could be constructed. This approach was implemented in the activated database MATRIX accumulating oligonucleotide frequency matrices of the functional DNA sites.
How to Run the Program
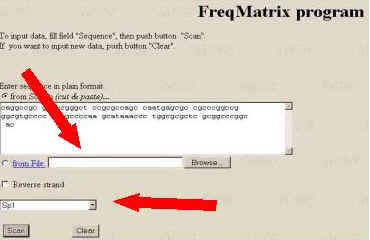
1. Input the DNA sequence into the field ‘Input DNA Sequence’. For example here given Human alpha-1 globin gene (EMBL HSHBA4; J00153), region (10379; 10478). This sequence containing the SP-1 transcription factor binding site (TRRD P00047).
2. Select one of transcription factor binding sites from the list and denote strand.

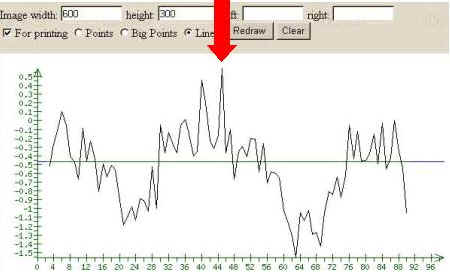
3. Start the tools processing by clicking the button ‘Scan’. The tools output represents the profile of the Score value. The positive peaks of this profile could point to the potential site recognized.
Ponomarenko M.P., Ponomarenko J.V., Frolov A.S., Podkolodnaya O.A., Vorobyev D.G., Kolchanov N.A., and Overton G.C. (1999) Oligonucleotide frequency matrices addressed to recognizing functional DNA sites. Bioinformatics, V. 15, N. 7/8, P. 631-643.