[EADER_SQ] [LEADER_KN] [LEADER_REF] [LEADER_SCI] [Paper] [Principles]
The LEADER_RNA program was designed to evaluate mRNA translational activity (Kochetov et al., 1999). It is based on significant differences between contextual features of mRNAs of eukaryotic genes expressed at high and low levels (Kochetov et al., 1998). The program applies these characteristic features to evaluate similarity of user-defined 5'UTR sequence with those of high- and low-expression genes. Currently, Leader_RNA contains data on three eukaryotic taxa including mammals, dicot and monocot plants.
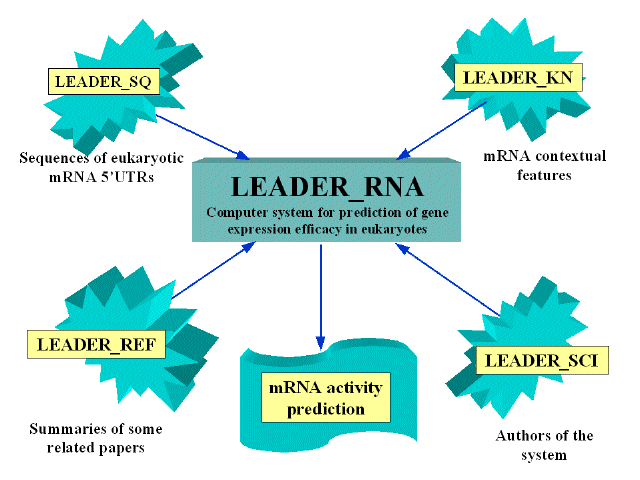
Knowledge base Leader_RNA is installed at the SRS platform. It contains five data files:
LEADER SQ accumulates the sequences of 5'UTRs of high- and low-expression mRNAs (used as the training samples for prediction program).
LEADER REF contains brief summaries of several papers concerning mRNA features influencing translational activity.
LEADER WHY contains brief description of contextual features of high and low expression mRNAs (stored in Leader_SEQ) that may be used for prediction of translation activity of user-defined mRNAs.
LEADER SCI contains the links displaying the authors of the computer system.
Basic principles of prediction program
© 1997-99, IC&G SB RAS, Laboratory of Theoretical Genetics