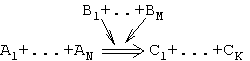 ,
, GeneNet: a database for gene networks and its automated visualization through the Internet
Kolpakov F.A., Ananko E.A., Kolesov G.B. and Kolchanov N.A.
Dagstuhl seminar “Bioinformatics” Modelling and simulation of gene regulation and metabolic pathways. 1998
The gene network concept. The physiological functions of organisms are accomplished through the coordinated regulation of the expression of a large number of genes. Hence, there exist complex networks: the gene ensembles functioning in a coordinated manner to provide vital functions, the fine regulation of physiological processes, and the responses to external stimuli.
The functional elements of a gene network are: (1) a gene ensemble interacting when certain biological functions are performed; (2) proteins encoded by these genes; (3) signal transduction pathways providing gene activation in response to an external stimulus; (4) a set of positive and negative feedbacks stabilizing the parameters of the gene network (autoregulation) or providing a transition to a new functional state; and (5) external signals, hormones, and metabolites that trigger the gene network or correct its operation in response to the changes in physiological parameters.
Databases on gene networks. Experimental data on the features of gene function have been rapidly accumulated during the last ten years resulting in development of several specialized databases. The major of them are (1) CSNDB, the Cell Signaling Networks Database (Igarashi and Kaminuma T, 1997; http://geo.nihs.go.jp/csndb.html); (2) SPAD, the Signaling Pathway Database; (3) BRITE, the Biomolecular Reaction Pathways for Information Transfer and Expression (http://www.genome.ad.jp/brite/brite.html); (4) KEGG, the Kyoto Encyclopedia of Genes and Genomes (Goto et al., 1997; http://www.genome.ad.jp/kegg/kegg.html); and (5) GeNet, the Gene Networks Data Base.
All the above databases contain manually drawn interactive diagrams of the signal transduction pathways and gene networks described. Automated construction of diagrams from the formalized information appears to be a promising direction. EcoCyc was the first convincing demonstration of the efficiency of automated diagram generation for metabolic pathways (Karp et al., 1996); however, the gene network databases available are not provided with such tools.
The GeneNet database.We have developed an object-oriented database GeneNet (Kolpakov et al., 1998), compiling the information on the gene networks of antiviral response and erythropoiesis regulation. The information contained in the databases IIG-TRRD (Anan'ko et al., 1997) and ESG-TRRD (Podkolodnaya, Stepanenko, 1997), respectively, was used for their formalized description.
A chemical formalism was employed as a basis for describing the events occurring in the gene network. Thus, any event is described as follows:
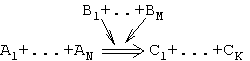 ,
,
where A is the objects entering into reaction; B, the objects affecting the course of reaction; and C, the products of reaction. Basing on this model, we consider two types of interactions: (1) reaction (indicated by double arrow in the scheme), that is, formation of a new object or acquisition of a new property by the object, and (2) regulatory event (ordinary arrow), that is, the effect of an object on certain reaction.
The following objects participating in the events are considered: (1) cells (tissues, organs); (2) genes; (3) proteins and protein complexes; and (4) nonprotein regulatory substances and metabolic products. In the GeneNet database, each object type is described in a separate table: (1) CELL, containing the information on cells, tissues, and organs; (2) PROTEIN, on proteins and protein complexes; (3) GENE, on genes and their regulation patterns ; (4) SUBSTANCE, on nonprotein regulatory substances and other metabolic products; (5) STATE, on physiological functions and the state of the organism ; (6) RELATION, on interrelationships of these objects; (7) SCHEME, containing the formalized description of the gene network graph that includes the list of objects forming this gene network, the list of their interrelationships, and directions for optimal layout of the objects in the graph; and (8) LITER, containing the references. The GeneNet has references to the databases EMBL, SWISS-PROT, TRRD, TRANSFAC, EPD, and MEDLINE.
Automated generation of gene network diagrams. The GeneNet database is designed to allow the automated construction of the gene network diagrams basing on their textual description. A specialized Java program was created for this aim. It is accessible via the Internet at http://wwwmgs.bionet.nsc.ru/mgs/systems/genenet//.
The gene network diagram is represented in a form of graph with nodes corresponding to various objects and edges, to interrelationships between these objects. Each component of the gene network has its own image reflecting the peculiarity of the object.
A clear-cut compartmentalization is characteristic of all biochemical reactions in the organism. Hence, the gene network is described at three levels: (1) organism level; (2) single cell level; and (3) single gene level. This allows us both to take into account that the components of a gene network can be scattered through different organs, tissues, cells, and cellular compartments and to describe different regulation levels of the gene network.
The system of filters. The table SCHEME contains a consolidated description of the gene network based on experimental data obtained in different species, cell types, and under different conditions. The default diagram is built basing on the entire table. However, the system of filters allows the user to select for graphical representation only those objects and their interrelationships that have been described experimentally in a specified species, cell type, and/or in response to a certain stimulus.
The work was supported by the Russian Foundation for Basic Research (grants Nos. 96-04-50006, 97-07-090309, 97-04-49740, and 98-04-49479), Russian National Program on Human Genome, and Russian Committee on Science and Technology. The authors are grateful to O.A. Podkolodnaya for kindly providing the information on regulation of erythropoiesis.
References
Anan’ko,E.A., Bazhan,S.I., Belova,O.E., and Kel',A.E. (1997) Mechanisms of transcription of the interferon-induced genes: a description in the IIG-TRRD information system. Mol. Biol. (Mosk.), 31, 592-605.
Podkolodnaya,O.A. and Stepanenko,I.L. (1997) Mechanisms of transcription regulation of the erythroid-specific genes. Mol. Biol. (Mosk.), 31, 671-683.
Karp,P, Riley,M., Paley,S., and Pellegrini-Toole,A. (1996) EcoCyc: Electronic encyclopedia of E. coli genes and metabolism. Nucleic Acids Res., 24, 32-40.
Kolpakov,F.A., Ananko,E.A., Kolesov,G.B., and Kolchanov,N.A. (1998) Bioinformatics, 14, in press.
Goto,S., Bono,H., Ogata,H., Gujibuchi,W., Nishioka,T., Sato,K., and Kanehisa,M. (1997) Organizing and computing metabolic pathways data in terms of binary relations. In: Proc. Pacific Symp. Biocomputing, 1997, 175-186.
Igarashi,T. and Kaminuma,T. (1997) Development of a cell signaling Network Database. In: Proc. Pacific Symp. Biocomputing, 1997, 187-197.