MATHEMATICAL MODEL OF ERITHROID CELL DIFFERENTIATION REGULATION
*Ratushny A.V., Podkolodnaya O.A., Ananko E.A., Likhoshvai V.A.
Institute of Cytology and Genetics SB RAS, Russia
e-mail: ratushny@bionet.nsc.ru
*Corresponding author,
Keywords: erythroid cell, differentiation, gene network, regulation, mathematical model, computer analysis
Resume
Motivation:
Construction of an adequate mathematical model is required for investigating possible function modes of the complex nonlinear erythroid cell differentiation and maturation gene network and determining optimal strategies of its regulation while solving particular problems, therapeutic application included.
Results:
A dynamic model of the function of gene network regulating erythroid cell differentiation is constructed. The model is described in terms of elementary processes—biochemical reactions. The optimal set of the model parameter values is determined. Dynamic behavior patterns of the system under different conditions are simulated numerically.
Introduction
The hematopoietic tissue belongs to self-regenerating systems of the organism, regulated and self-regulated by specific patterns. Maintenance of a definite volume of blood cells is one of the necessary conditions of the organism function. From this standpoint, the insight into cell proliferation and differentiation of the hematopoietic tissue provided by a theoretical study is of both basic and biomedical importance. A dynamic model of the function of gene network regulating erythroid cell differentiation is constructed in the work. The model is described in terms of elementary processes—biochemical reactions. Numerical experiments are used to determine the set of parameters allowing the calculations to comply with the published experimental data. Dynamic behavior patterns of the system under different conditions are simulated numerically. The results obtained are compared with the relevant experimental data.
Regulation of erythroid cell differentiation
The main stages of erythroid cell differentiation are qualitatively represented in the GeneNet database (http://wwwmgs.bionet.nsc.ru/mgs/systems/genenet/). The hormone erythropoietin interacts with the receptors of immature erythroid cells (erythroid stem progenitors of CFUe type) and stimulates their proliferation, hemoglobin synthesis, and synthesis of the enzymes involved in heme biosynthesis, that is, maturation and differentiation of erythroid progenitors [Podkolodnaya et al., 2000]. A low partial pressure of venous oxygen (hypoxia) stimulates erythropoietin synthesis.
The system regulating the erythroid cell differentiation displays a pronounced positive feedback. Erythropoietin interaction with the cellular receptor activates GATA-1 transcription factor, a key regulator of erythrocyte differentiation. GATA-1 stimulates the syntheses of a and b globins as well as the heme-synthesizing enzymes. In addition, GATA-1 activates its own gene and the gene coding for erythropoietin receptor (positive feedback). Hem, a , and b globins form hemoglobin, the major component of the mature erythrocyte.
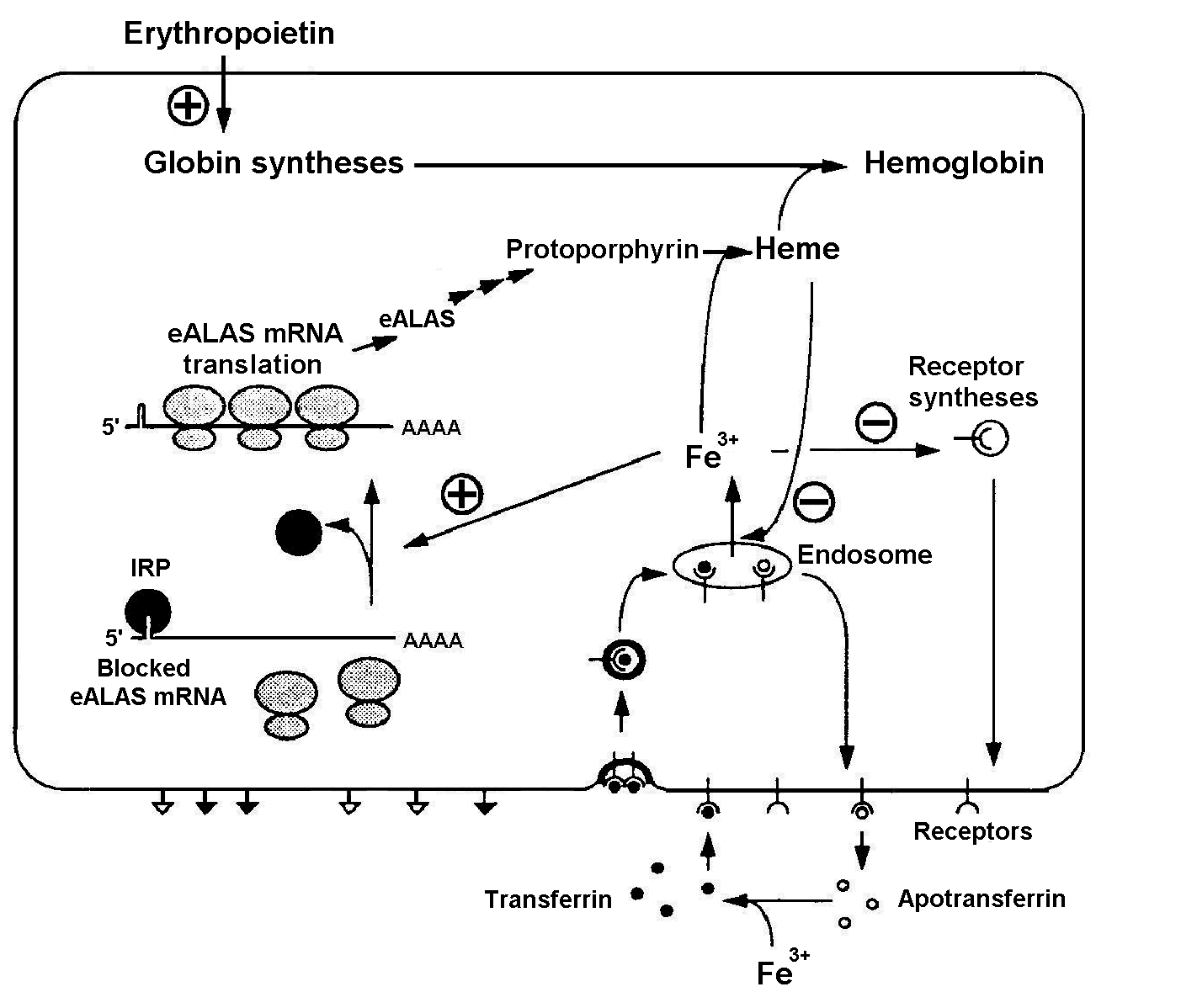
Fe3+ ions are necessary for heme synthesis and, respectively, hemoglobin synthesis. Iron regulating protein (IRP) plays an important role in heme biosynthesis [Kuhn, 1994]. The system regulating influx of Fe3+ ions into the erythroid cell is shown in Fig. 1. It is evident from the scheme that it has not only positive, but also negative feedbacks. The specific blood serum protein transferrin transports iron from gastric mucosa and spleen sinus parenchyma into bone marrow [Fedorov, 1976]. Transferrin interacts with the membrane receptors and enters the cell through endocytosis. In the absence of Fe3+ ions, IRP binds the mRNA of a heme-synthesizing enzyme, 5-aminolevulinate synthetase (eALAS), reducing the synthesis. eALAS is completely unbound when the Fe3+ concentration in cell is high. Decrease in the globin syntheses due to certain reasons, resulting in an excess heme concentration, may inhibit the influx of Fe3+ either directly or indirectly through decrease in the synthesis of transferrin receptors (negative feedback) [Kuhn, 1994]. An excess of heme also has a positive effect on globin syntheses [Ponka, 1997].
Figure 1. Regulation of Fe3+ influx to the erythroid cell and the control function of iron regulating protein (IRP): eALAS, erythroid 5-aminolevulinate synthetase [Kuhn, 1994].
Methods and algorithms
The system was described and analyzed by methods detailed in [Ratushny et al., 2000].
Mathematical model
The system regulating the erythroid cell differentiation is described with 119 kinetic blocks. The model contains 68 dynamic variables and 178 reaction constants. Values of the parameters were determined in different ways. Experimental data, partially listed in table below, were used to determine a number of enzymatic kinetic parameters of this system.
Table. Some reaction constants.
Enzyme |
Substrate |
Organism |
Organ |
Kc, sec- 1 |
Km, mM |
| Aminolevulinate synthase | Glycine Succinyl-CoA |
Rattus norvegicus [Scholnick, 1972] | Liver | (-) (-) |
11.0 0.07 |
| Aminolevulinate synthase | Glycine Succinyl -CoA |
Mus musculus [Tan & Ferreira, 1996] | Liver | 2.05e-02 2.05e-02 |
8.39 1.82 |
| Aminolevulinate synthase | Glycine Succinyl -CoA |
Saccharomyces cerevisiae [Volland & Felix 1984] | 7.17e+01 7.17e+01 |
3.0 0.002 |
|
| Porphobilinogen synthase | 5-aminolevulinate | Homo sapiens [Gibbs, 1985] | Erythrocyte | 11.67 |
0.29 |
| Porphobilinogen deaminase | Porphobilinogen | Homo sapiens [Smythe & Williams, 1988] | Erythrocyte | 2.93e-2 |
(-) |
| Porphobilinogen deaminase | Porphobilinogen with URPIII synthase | Homo sapiens [Frydman & Feinstein, 1974] | Erythrocyte | (-) |
0.13 0.051 |
| Porphobilinogen deaminase | Porphobilinogen | Rattus norvegicus [Mazzetti & Tomio, 1988] | Liver | 3.43e-4 |
0.017 |
| Uroporphyrinogen decarboxylase | Heptacarboxyl porphyrinogen | Homo sapiens [Mukerji & Pimstone, 1987] | Erythrocyte | 1.11e-3 (9.13e-4) |
2.31e-3 (7.1e-4) |
| Uroporphyrinogen decarboxylase | Porphyrinogen I Porphyrinogen III |
Saccharomyces cerevisiae [Felix & Brouillet, 1990] | 2.39 0.906 |
1.0e-5 6.0e-6 |
|
| Coproporphyrinogen oxidase | Coproporphyrinogen III | Bos taurus [Yoshinaga & Sano, 1980] | Liver | 0.203 (0.178) |
0.048 (0.025) |
| Coproporphyrinogen oxidase | Coproporphyrinogen III | Mus musculus [Kohno et al., 1996] | Liver | 0.291 |
0.047 |
| Protoporphyrinogen oxidase | Protoporphyrinogen IX | Mus musculus [Dailey & Karr, 1987] | Liver | 2.49 |
5.6e-3 |
| Protoporphyrinogen oxidase | Protoporphyrinogen IX | Homo sapiens [Dailey & Dailey, 1997] | Placenta | 1.75e-1 |
1.7e-3 |
| Protoporphyrinogen oxidase | Protoporphyrinogen IX | Bos taurus [Siepker, 1987] | Liver | 6.51 |
1.66e-2 |
| Ferrochelatase | Protoporphyrin Fe citrate |
Bos taurus [Nakahashi, 1990] | Liver | 7.77e-1 7.77e-1 |
1.27e-2 3.51e-3 |
| Ferrochelatase | Protoporphyrin Fe citrate |
Rattus norvegicus [Taketani, 1981] | Liver | 0.63 0.63 |
2.85e-2 3.74e-2 |
The values of model parameters lacking in the literature were verified through numerical experiments. At this stage, the values of parameters were selected so that the integral system behavior would comply maximally with the available experimental data on the dynamic characteristics of the system’s behavior, including the following:
Hemoglobin is actively accumulated starting from the stage of basophilic erythroblast. Hemoglobin and other components are usually synthesized during G1 and the beginning of S phases of DNA synthesis [Kozinets & Goldberg, 1982]. The total (transit) time from proerythroblast to reticulocyte formation is 120 h (5 days). Two fifth of the overall cell cycle occur during G1 and the beginning of S phases [Fedorov, 1976]. Thus, the overall syntheses of internal components in a differentiating erythroid cell require approximately 50 h.
The number of transferrin receptors on the surface of an immature erythrocyte amounts to approximately 104 to 105 per cell; they are not detected in the progenitor cell [Kuhn, 1994].
The rates of hemoglobin synthesis in the cells at the stages of proerythroblasts and basophilic erythroblasts equal approximately 0.5 pg/h, that is, about 4.7*106 molecules/h for an average cell with a diameter of 10 m m; at the stage of reticulocyte, the rate decreases fivefold, that is, to about 106 molecules/h [Kozinets & Goldberg, 1982].
Each erythrocyte contains about 280 million hemoglobin molecules [Fedorov, 1976].
Theoretical numerical calculations according to this model demonstrate the following:
Oscillating synthesis dynamics are typical of heme, a and b globins, and the components of this molecular genetic system associated with the IRP control function and involved into Fe3+ influx into the erythroid cell (Fig.2, b,c,d,e). This is due to the network of negative and positive feedbacks, shown in Fig. 1.
No excess of free heme is accumulated in the differentiating erythroid cell (Fig.2, f). First, it is due to control of heme biosynthesis by IRP, which binds eALAS mRNA, and eALAS is, in turn, the enzyme involved in heme synthesis at the initial stage. Second, the excess of heme has a positive effect on globin biosyntheses.
The number of transferrin receptors on the surface of an immature erythrocyte reaches approximately 104 per cell, whereas these receptors are initially absent in the progenitor cell (Fig.2, c).
The rate of hemoglobin synthesis several hours after the beginning of erythropoietin effect equals approximately 5*106 molecules/h; maximum (8*106) is observed at 31 hours; in ten hours after the genes are switching off, approximately 106 molecules/h. Note that after 25 hours, the rate of synthesis demonstrates oscillating dinamics (calculations not shown).
An erythrocyte contains about 280 million molecules of hemoglobin on cessation of its synthesis at 74 hours (Fig. 2, d).
Figure 2. Dynamics of the main components on the system regulating the erythroid cell differentiation. By TfRTf are denoted transferrin receptors bound to transferrin (calculations by a model).
Acknowledgements
The authors are grateful to Galina Chirikova for translation of the manuscript into English and N.A. Kolchanov for fruitful discussions. The work was supported by National Russian Program "Human Genome" (No 106), Integrational Science Project of SB RAS "Modelling of basic genetical processes and systems".
References