[Aim] [Background] [Program description] [Implementation] [Limitations] [Acknowledgements] [Contacts]
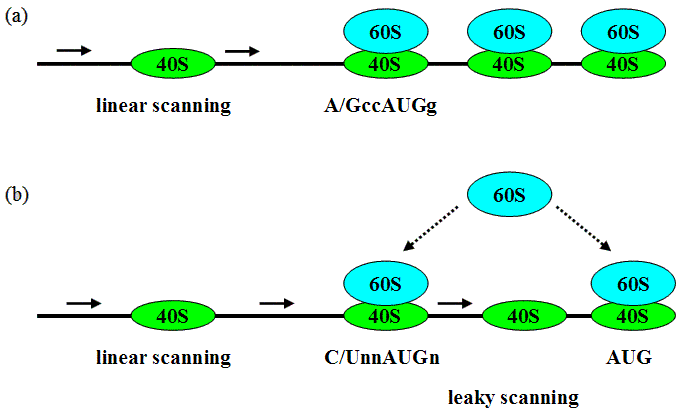
Translation of most eukaryotic mRNAs is likely to be initiated by a linear scanning mechanism (Kozak, 2002). According to the scanning model, 40S ribosomal subunits can either initiate translation at the proximal AUG codon in a suboptimal context or miss it and initiate translation at downstream AUG(s) (Fig.1). It is assumed that recognition of the translation start codon by eukaryotic ribosomes depends on its nucleotide context. For mammalian and plant mRNAs, the most crucial elements of AUG context are a purine at position -3 and a guanine at position +4 (PuNNAUGG; Kozak, 2002).

Fig.1. Linear scanning model of eukaryotic mRNA translation: (a) 40S ribosomal subunits bind to the capped 5' mRNA end and move linearly until the proximal AUG codon in optimal context, whereupon ribosomal subunits associate and translation elongation begins. (b) Recognition of AUG triplet as translation start codon depends on its nucleotide context: if a proximal AUG is located in a suboptimal context, some 40S ribosomal subunits can miss it, continue scanning, and initiate translation at downstream AUG(s) ("leaky scanning").
One might expect that mRNA should possess the features providing efficient translation, including recognition of a genuine translation start site (TSS). However, the fraction of eukaryotic mRNAs with the start AUG codon in a suboptimal context is relatively large (Rogozin et al., 2001). It is likely that at least some mRNAs with a suboptimal start codon context contain other signals providing additional information for efficient TSS recognition. It has been reported earlier that stable hairpins precisely positioned downstream of an AUG codon in a suboptimal context can increase the efficiency of its recognition as a translation start site (Kozak M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc. Natl. Acad. Sci. USA 1990. 87. 8301-8305). The hairpin was placed at the distances of 5, 11, 17, and 35 nucleotides downstream of the CDS beginning (Fig. 2). It was reported that the hairpin located at the distances of either 5 or 35 nucleotides did not increase AUG recognition. However, the hairpin located at a distance of 17 or 11 (to a lesser extent) nucleotides increased translation initiation level considerably.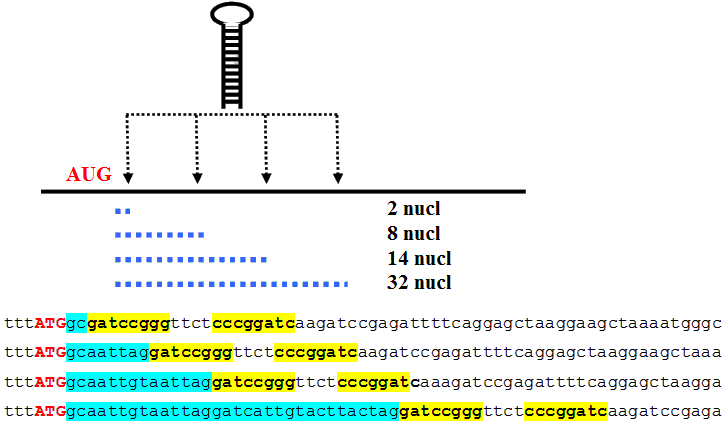
Fig. 2. Description of the experiment reported by Kozak (1990): The hairpin (marked with bold type and yellow boxes) was placed at the distances of 5, 11, 17, and 35 nucleotides downstream of the CDS beginning (marked with blue boxes).
It was assumed that the hairpin located at the distance of ca. 17 nucleotides from the CDS beginning could slow down the movement of 40S ribosomal subunits along mRNA in a position providing an efficient interaction between the met-tRNAi-located anticodon and the start AUG codon (Fig.3).

Fig. 3. Putative role of downstream secondary structure in the recognition of a translation start codon in a suboptimal context: it was hypothesized that a stable hairpin located 12-15 nucleotides downstream of AUG can delay the movement of 40S ribosomal subunit at a point where [AUG codon : UAC anticodon] complementary interaction occurs at a maximal efficiency. Such a delay increases the probability of recognition of AUG codon in a "weak" context as the translation start site (Kozak, 1990).
According to the experimental data (Kozak, 1990), the hairpins started either upstream or downstream of certain “critical” region did not compensate for the start codon “weakness”. In particular, continuous secondary structure started at 5th nucleotide of coding sequence did not increase translation initiation efficiency despite it included the critical 11th and 17th positions (Kozak, 1990). Basing on this observation, AUG_hairpin program predicts the stem-loop structures whose 5’-borders are located within the critical region (from 12th to 18th nucleotides by default). An appropriate stem-loop structure can also be a part of a more complex secondary structure started upstream of the critical region. In this case an eligible hairpin has to be separated from upstream secondary structure elements by some impaired segment (e.g., loop) (Fig. 4). We assumed that the 40S ribosomal subunit moving from the 5’-end of mRNA could pause successively at each stable stem of a complex stem-loop structure waiting them to melt. Basing on the Kozak’s experiment, we assumed that the 40S ribosomal subunit moving from the 5’-end of mRNA could pause successively at each stable stem of a complex stem-loop structure waiting for them to melt. If such pausing occurs in a proper place, the recognition of a “weak” start codon is facilitated.
We prepared the on-line program (AUG_hairpin) for the prediction of a potential secondary structure compensating for a suboptimal context of the translation start site. It contains the program foldRNA from Vienna RNA package v.1.4, implemented as subroutine for prediction of RNA secondary structure. The program also provides the user with an opportunity to define the borders of a critical segment where the occurrence of a stable hairpin can improve AUG recognition. The program was written in C++ and runs in a Unix environment. Algorithm consists of the following main steps:
-- Prediction of RNA optimal secondary structure for 5’-UTR-CDS fragment
-- Analysis of a secondary structure by foldRNA;
-- Checking the occurrence of a stem-loop structure located a certain distance downstream of an AUG codon (12 to 18 nucleotides by default, user can change this range (minimal value for starting position is 1 (i.e., a hairpin located immediately at the CDS beginning)). The program also takes into account the following criteria:
a) To be taken into consideration, the hairpin should be separated from other upstream helices by a loop including at least 2 nucleotides.
b) The background experiments have been done with perfect hairpin(s) (Kozak, 1990). Conventionally a stem is perfect when it does not contain any interrupting loops; an imperfect stem includes short mismatches (one-nucleotide bulges or 1+1 inner loops) which presumably do not interrupt stacking interactions. AUG_hairpin provides the user with an opportunity to take into account either only perfect hairpins or the hairpins containing some mismatches (bulge or interior loop size may be adjusted; Fig. 5.).
-- Display html-page, containing results of calculation: energy of the whole secondary structure, position of a stem-loop structure and total energy of helices composing stem-loop; Secondary structure of the analyzed mRNA fragment is visualized. Start AUG position is marked in red; the borders of a critical RNA segment are marked in green; nucleotides composing an eligible hairpin, in blue.
1. Input 5'-UTR and CDS sequences. 10 nucleotides of 5'-UTR lying immediately upstream the start AUG codon and ca. 100 5'-end nucleotides of CDS (starting from AUG codon) are enough for calculation (larger sequences will be truncated). Note: only a,t(u),g,c symbols may be used.
2. Select parameters of eligible hairpins:
- select the critical region where the hairpin's 5'-border should be located (from 12th to 18th nucleotides by default)
- select only perfect hairpins or the hairpins with some mismatches (in the latter case the size of bulges or interior loops (1 nucleotide by default) may be adjusted)
3. Program report contains the figure of the predicted secondary structure, its energy, position of the stem-loop structure and total energy of helices composing the eligible stem-loop.




Fig. 4. Prediction of putative secondary structure in Kozak's experimental sequences (Kozak, 1990). Hairpins located over the distance from 12 to 18 nucleotides were taken into account. Each fragment contains:
- sequence (start codon is marked in red, the distance between ATG and the hairpin is marked
in blue, inverted repeats forming the hairpin are bolded and marked in yellow).
- hairpin in Kozak’s notion (start codon is marked in red, secondary structure is marked
in blue);
- secondary structure predicted by AUG_hairpin (start codon is marked in red, area boundaries (12 to 18) are marked
in green and secondary structure, in blue)

Fig. 5. An example of imperfect hairpin eligible by default (an imperfect stem includes short mismatches (one-nucleotide bulges or 1+1 inner loops which presumably do not interrupt stacking interactions). User may change these parameters (i.e., AUG_hairpin will select only perfect hairpins or imperfect hairpins with the user-defined maximal loop size).
- translation initiation mechanisms
Translation of most eukaryotic mRNAs is likely to be initiated by a linear scanning mechanism, but some alternative mechanisms are also possible (e.g., internal ribosome entry sites, ribosome shunts, etc.). However, even though 40S ribosomal subunits bind to an internal 5’UTR segment, further they can scan nucleotide sequence linearly in 3’-direction until the translational start codon. Therefore, such a compensatory hairpin can potentially improve translation of some IRES-containing mRNAs with a translation start codon in a suboptimal context.
- program parameters
Program takes into account only local stem-loop structures located at a certain distance downstream of AUG codon within a restricted mRNA segment. It is possible that mRNA folds in a complex structure and a predicted “compensatory hairpin” can be a part of an extended secondary structure. Program also does not take into account higher order structures or distant interactions. Program cannot take into account protein-RNA interactions.
Alex V. Kochetov ak@bionet.nsc.ru
Andrey Palyanov palyanov@academ.org