The program is designed for estimation of symbolic data compression.
The sequence to be analyzed should be entered into the text-box in the FASTA-format.
Let us consider the task of searching for context dependencies in the set of AP-1 binding sites (32 kB, you can download these sequences from Eukaryotic Transcription Factor Binding Sites Compilation - http://wwwmgs.bionet.nsc.ru/mgs/dbases/nsamples/).
This set in a FASTA-format is entered into the input text box.
Set all other parameters by default. (More detailed description of the program is given in Help page).
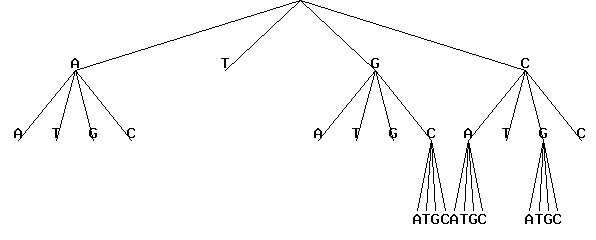
Press button "Execute". The resulting picture in GIF-format is shown in the bottom of this screen.
The results of the program execution will be displayed in new HTML-page as the figure shown:

If you uncheck parameter for picture:
Graphic output
and press "Execute" again, he results of the program execution will be displayed in new HTML-page in text format as given below.
The results contains user-defined parameters for the program, the same source tree as in picture in text format and all selected contexts (oligonucleotides) with their frequencies:
This output can be used for further prediction of DNA functional sites by the VMM program
(just cut and paste text in the corresponding window of model parameters)
Set context len:3 Set: Show all statistics Set legend for alphabet: Set model: Rissanen +0.5 pseudo count model >AP_10001 -------------- Data information -------------- Alphabet size:4 Alphabet contains:ATGC Context lenght:3 Number of contextes:64 Complexity :15976.343353 This is tree in pseudographics (0-absent, 1-present) /treebegin 4 ATGC 1 1111 1111000011111111 0000000000000000000000000000000000000000000011111111000011110000 (File size without comments) Length=8142+( 2) Standard compressed file size=2035.5000 Complexity[0][0]=15976.3434 Direct calculation=16243.1322 (Compression = 0.9836) Entropy=-SUM p*log(p) =1.9927 (*n=16224.3197) Full context tree complexity (3 levels)=16049.4487 (Compression relative to full tree=0.9954) +86.492+90.015+303.119+212.223+177.096+88.669+213.017+228.685+292.880+355.262+366.817+290.590+940.399+563.361+973.922+1082.446+1166.418+1243.845+1452.686+914.056+1272.908+3661.437 Control complexity sum of leaves 15976.3434 == 15976.3434 --Control complexity sum of leaves 0.0000 == 15976.3434 Context - Variability - Complexity /contextbegin GCAA= 24 GCAT= 19 GCAG= 56 GCAC= 22 ----------GCA=129 24 19 56 22 sum=121 ( 86.4921) ( -1.2781) GCTA= 24 GCTT= 49 GCTG= 60 GCTC= 29 ----------GCT=161 24 49 60 29 sum=162 ( 90.0154) ( -1.3204) GCGA= 22 GCGT= 20 GCGG= 59 GCGC= 59
. . .
----------CC=642 149 161 122 206 sum=638 ( 1272.9083) ( -1.3685) --Length is 2.Leaves found:9 TA=287 TT=474 TG=616 TC=491 ----------T=1861 287 474 616 491 sum=1868 ( 3661.4369) ( -1.3528) --Length is 1.Leaves found:1 ---Total number of leaves 22 ---Simple empirics. Complexity of the tree relative to the full 3-tree is 0.085938 ---- Context Length. Leaves found ------- --- 1 1 --- 2 9 --- 3 12 --- 4 0
This resource has been developed in Institute of Cytology and
Genetics, Novosibirsk, Russia
Authors: Yu.L.Orlov, V.P.Filippov, V.N.Potapov
Contributor: S.V.Lavryushev, D.A.Grigorovich
Leader: N.A.Kolchanov
The research was partially supported by the Russian Foundation for Basic Research (RFBR)