CRASP |
Pairwise
positional correlation estimation results.
[Tutorial page] |
Comments: The main calculation parameters are given below.
MULTIPLE ALIGNMENT OF "HMEN_DROME_455
" AND HOMOLOGOUS SEQUENCES.
NUMBER OF SEQUENCES: 372.
ALIGNMENT LENGTH: 74.
AMINO ACID PHYSICO-CHEMICAL CHARACTERISTICS: Isoelectric point (Zimmerman et al., 1968).
REFERENCE: J. Theor. Biol. (1968) v.21, p.170-201.
WEIGHTS: according to Felsenstein(1985),Am.Nat.,125,1-15.
Comments: The
main parameters fro the correlation matrix: its type, size, and the
critical value of the correlatiojn coefficient at
a given significance level are given below.
MATRIX TYPE: Partial correlation coefficient matrix.
NUMBER OF ANALYSED POSITIONS: 47. Crit.Value (99.9%): rc=0.182170
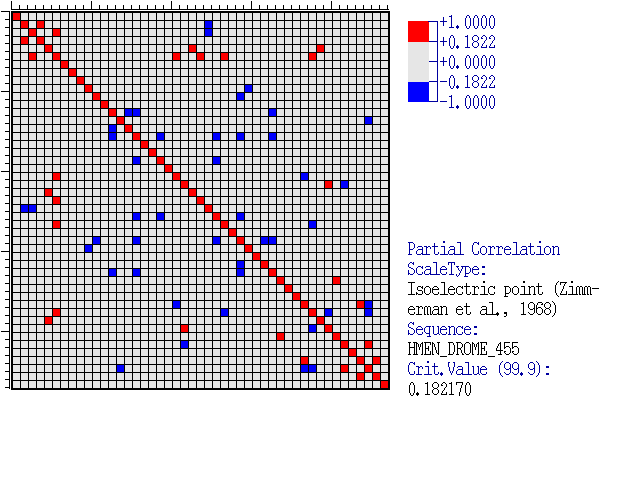
Comments: The
diagram for the correlation coefficient matrix is as follow. Depending on
the output options, the matrix can be in a textual format, an HTML table,
a color diagram. For the latter option the indexing of alignment
positions are plotted if the diagram accommodate them.
Pairwise dependencies matrix representation.
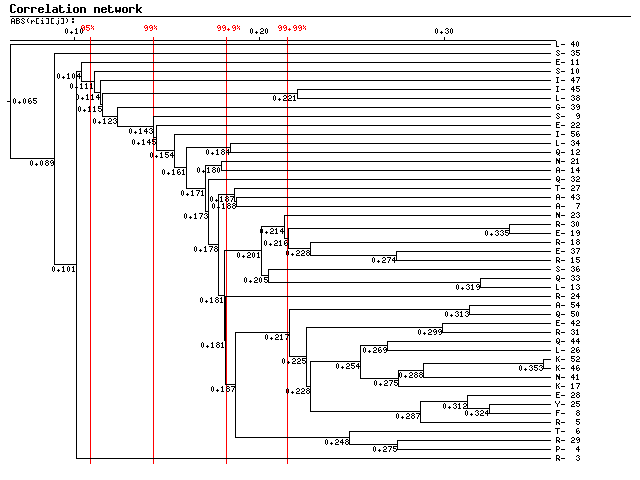
 Comments: Below is given a diagram for the hierarchical clustering of the alignment
positions based on their mutual correlation coefficient. Vertical red lines designate
the critical values for the correlation coefficient at different significance levels.
The position designations are given at the left.
Binary tree diagram for clusters of correlated positions.
Comments: Below is given a diagram for the hierarchical clustering of the alignment
positions based on their mutual correlation coefficient. Vertical red lines designate
the critical values for the correlation coefficient at different significance levels.
The position designations are given at the left.
Binary tree diagram for clusters of correlated positions.
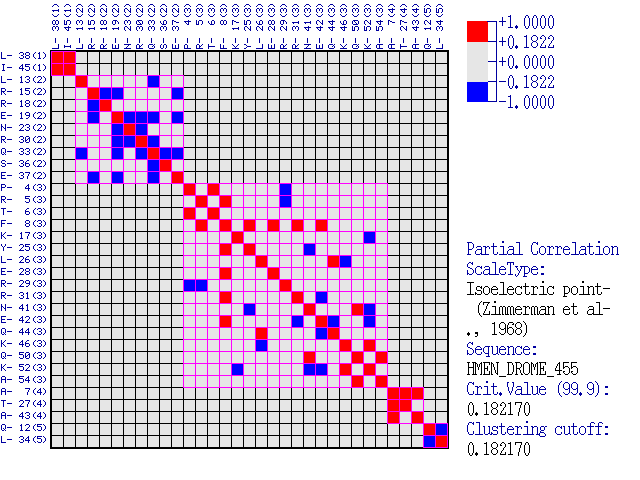
 Comments: Below is given a submatrix for the correlation coefficient for clusters
that result from cutting off the branches of above tree ath the correlation
coefficient value rcut-off.
Close view of correlations in significant clusters.
Comments: Below is given a submatrix for the correlation coefficient for clusters
that result from cutting off the branches of above tree ath the correlation
coefficient value rcut-off.
Close view of correlations in significant clusters.
 Comments: Below is given a hypertext reference to file with the means and
variations in the physicochemical property values for each alignment position.
Positional data.
Comments: Below is given a hypertext reference to file with the means and
variations in the physicochemical property values for each alignment position.
Positional data.
Comments: Below is given a hypertext reference to the file with the frequencies of
occurrence of different amino acid types at alignment positions.
AminoAcid occurences at alignment positions.
Comments: Below is given a hypertext reference to the file with the correlation
matrix elements distribution and its statistical parameters.
Matrix elements distribution.
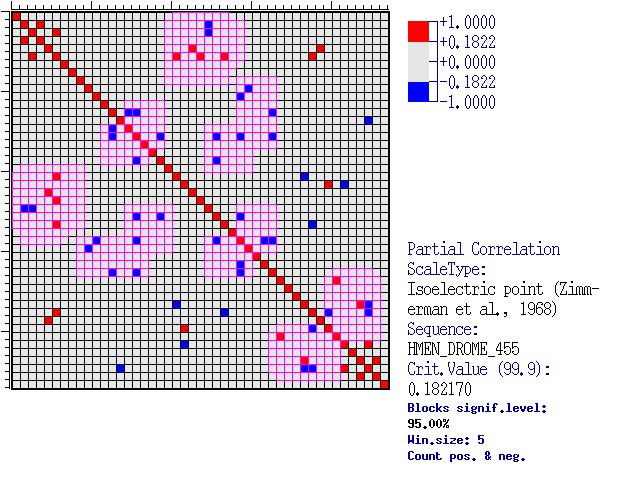
Comments: Below is given the diagram for correlation coefficient matrix,
the regions with predominantly significant correlation coefficients are highlighted in magenta.
Regions of high density of correlated pairs.
 Comments: Below is given a diagram for the hierarchical clustering of the alignment
positions based on their mutual correlation coefficient. Vertical red lines designate
the critical values for the correlation coefficient at different significance levels.
The position designations are given at the left.
Binary tree diagram for clusters of correlated positions.
Comments: Below is given a diagram for the hierarchical clustering of the alignment
positions based on their mutual correlation coefficient. Vertical red lines designate
the critical values for the correlation coefficient at different significance levels.
The position designations are given at the left.
Binary tree diagram for clusters of correlated positions.
 Comments: Below is given a submatrix for the correlation coefficient for clusters
that result from cutting off the branches of above tree ath the correlation
coefficient value rcut-off.
Close view of correlations in significant clusters.
Comments: Below is given a submatrix for the correlation coefficient for clusters
that result from cutting off the branches of above tree ath the correlation
coefficient value rcut-off.
Close view of correlations in significant clusters.
 Comments: Below is given a hypertext reference to file with the means and
variations in the physicochemical property values for each alignment position.
Positional data.
Comments: Below is given a hypertext reference to file with the means and
variations in the physicochemical property values for each alignment position.
Positional data.
