Ab initio human miRNA prediction
Program description: The program for ab initio human miRNA prediction in an arbitrary genomic sequence.
Biological task that could be solved: Identification of novel human miRNAs.
Data input: Into the text-box (red arrow #1 in Fig.1), enter or insert from the clipboard a genomic sequence to be analyzed. You may input this sequence in the fasta format or in a plain text format without the comment line. The size limit is 10 000 symbols. Use the alphabets ATGC(atgc) or AUGC(augc), any other symbol in the nucleotide sequence will give an error. Line foldings and spaces are ignored.
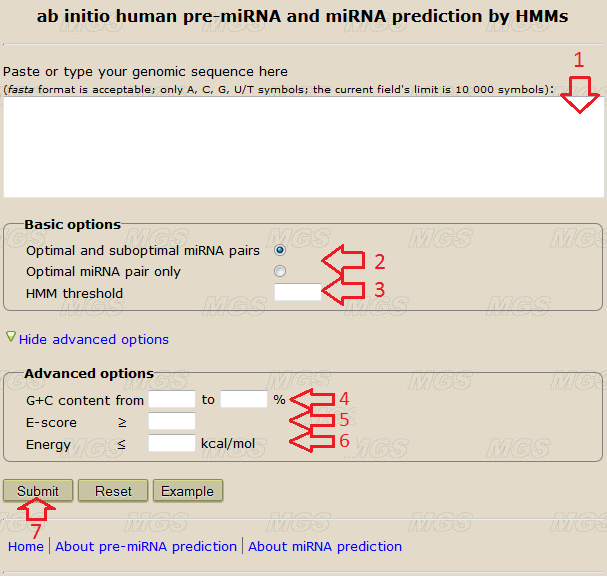
Figure 1. The main program window.
Program options: To predict miRNA/miRNA* duplex in the known miRNA precursor you need to input the pre-miRNA into the text-box (red arrow #1) and select "optimal miRNA pair" or "optimal and next suboptimal miRNA pairs" radio button (red arrow #2). Leave the other fields (red arrows #3-6) empty. The "optimal miRNA pair" option limits search to miRNA/miRNA* duplex with the highest score; the "optimal and next suboptimal miRNA pairs" limits search to the two top miRNA/miRNA* candidates with the highest scores. The result from the first option includes the result from the second option. If you need to predict miRNAs in an arbitrary genomic sequence you need additionally to adjust pre-miRNA prediction options (red arrows #3-6, see here).
Program execution: Start the program by clicking the button "Submit" (red arrow #7).
Data output: Program execution will bring up the resulting window with the data output.
They include the following (see the picture below):
• The number of the predicted pre-miRNA (red arrow #1);
• The start position of this pre-miRNA in the genomic sequence (red arrow #2);
• The pre-miRNA's stem-loop with marked miRNA/miRNA* duplex for the optimal miRNA/miRNA* duplex (red arrow #3) or for the best two variants (red arrows #3-4). The miRNAs are marked by the uppercase letters.
• The sequences of the predicted miRNAs.
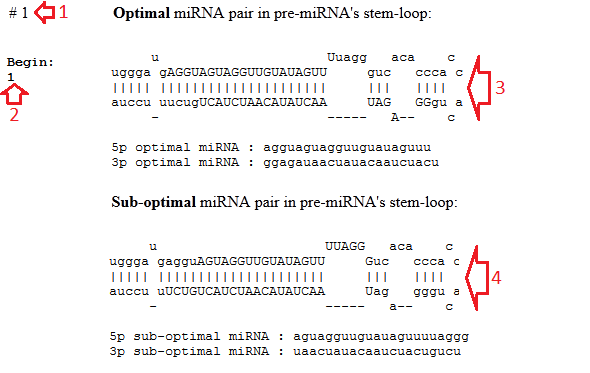
Figure 2. The resulting window.
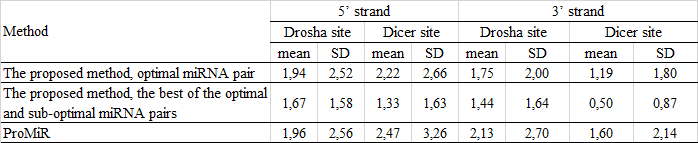
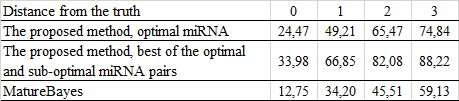
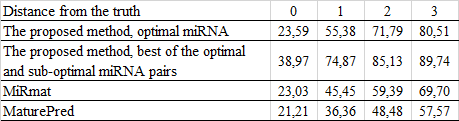
Evaluation of the human miRNA prediction:
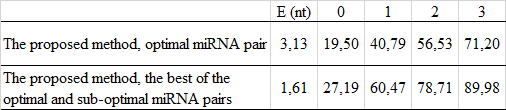
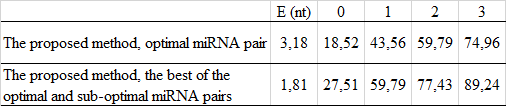
Comparing the efficiency with other programs for miRNA prediction:
RERERENCES:
MiRmat: He C, Li YX, Zhang G, Gu Z, Yang R, Li J, Lu ZJ, Zhou ZH, Zhang C, Wang J: MiRmat: mature microRNA sequence prediction. PLoS One 2012, 7(12):e51673.
maturePred: Ping Xuan, Maozu Guo, Yangchao Huang, Wenbin Li, Yufei Huang (2011) MaturePred: Efficient Identification of MicroRNAs within Novel Plant Pre-miRNAs. PLoS ONE, 6(11): e27422.
matureBayes: Gkirtzou K, Tsamardinos I, Tsakalides P, Poirazi P (2010) MatureBayes: A Probabilistic Algorithm for Identifying the Mature miRNA within Novel Precursors. PLoS ONE 5(8): e11843.
ProMiR: Nam, 2005. Nam J.W., Shin K.R., Han J., Lee Y., Kim V.N., Zhang B.T. 2005. Human microRNA prediction through a probabilistic co-learning model of sequence and structure. Nucleic Acids Res. 33(11), 3570–3581.