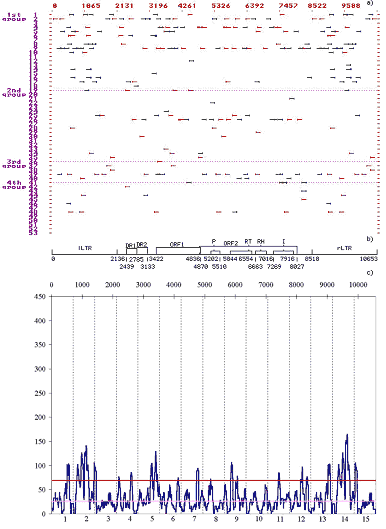
Designations. On an axes abscissas - sequence DNA mobile element in bp; on an ordinate axis - ascribed numbers of the regulatory sites (their titles are reduced in tab.1); 1 group - the sites of replication and transcription initiation and termination; 2 group - enhancers and silencers of chromosomal, viral, etc., genes; 3 group - the sites recognized by cellular protein transcription and translation factors; and 4 group - the sites recognized by protein receptors for inductive signals; the arrows mark the discovered location of regulatory sites on the left-directed (leftward arrows) and right-directed (rightward arrows) DNA strands.
(b) Schematic structure of the DNA sequence of retrotransposon Ulysses.
Designations. LTR, long terminal repeats; ORF1 and ORF2, big open reading frame; DR1 and DR2, direct repeats; P is the motif of amino acid sequence of protease domain; RT, of reverse transcriptase domain; RH, of RNase H domain; and I, of integrase domain.
(c) Consolidated distribution of the revealed motifs of functional sites along the Ulysses DNA sequence.
Designations. On an axes abscissas - number of segments of genome each size 1/15 length given MGE; on an ordinate axis - total number of nucleotides contained in the motifs of functional sites and falling within the scanning window by a size 75 bp. The upper direct line - 95%th level of the nonrandomness of the condensations of the motifs of functional sites; the lower direct line - average for 50 of casual sequences the same lengths and same nucleotide's of structure as DNA Ulysses distribution of motives functional saites.
Jamming of the motifs correlates with the potential regulatory regions in the LTR and the vicinity of the beginnings of the ORF2 and domains of the ORF2.