

 |
 |
| HOME | SITE MAP | GENENET DATABASE | PLANT-TRRD | PLANT ONTOLOGY |
| Photomorphogenesis Gene Network | Nodulation Gene Network | Flower development Gene Network |
Photomorphogenesis Gene NetworkThe Gene Network of the Photomorphogenesis is available via the Internet Olga G. Smirnova e-mail: planta@bionet.nsc.ru | |||
|
Light is the main regulator of plant development. Seedling photomorphogenesis is a program of switching from dark-adapted (skotomorphogenic) to light-adapted (photomorphogenic) development. Successful realization of this program ensures correct formation of vegetative and generative plant organs in future. The Photomorphogenesis gene network is used to accumulate information on functioning of plant genes, proteins and substrates providing plant light development. |

The Photomorphogenesis gene network. | ||
|
Using a system of filters, we can isolate local gene networks. The local gene networks regulate functioning of a repression complex in darkness, deactivation of this complex in light, activation of HY5 transcription factor and formation of a photomorphogenic response in the form of inhibition of hypocotyl growth, development of chloroplasts, and anthocyanin accumulation. The interactions among the elements of different local gene networks induce a hierarchical structure of the Photomorphogenesis gene network with COP1 as a main repressor and HY5 as a main positive regulator of seedling development. |
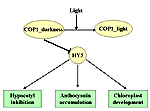
The structure of Photomorphogenesis gene network is presented in the form of local gene networks interactions. | ||
|
COP1 repression complex in darkness. In an etiolated seedling, light-activated nuclear (cab, rbcS, chs, cip4, cip7) and chloroplast (rbcL) genes are repressed by a complex with COP1 protein as the main component . A basal level of COP1 activity in darkness is provided by the cytoplasmic protein FIN219 and the IMPalpha1b importin. In the nucleus, COP1 affects ubiquitination and degradation of the transcription factor HY5, which plays an important role in the activation of gene transcription in light.COP1 represses chs, cip4, rbcS, cab genes and promotes degradation of the transcription factor HY5 in darkness. On the other hand, the activation of phyA gene transcription by COP1 leads to the high level of inactive photoreceptors in the cytoplasm. |
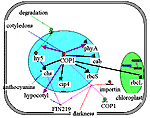
Application of "COP1_darkness" filter to the Photomorphogenesis gene network.
COP1 represses chs, cip4, rbcS, cab genes and promotes degradation of the
transcription factor HY5 in darkness. On the other hand, the activation of
phyA gene transcription by COP1 leads to the high level of inactive
photoreceptors in the cytoplasm. Black lines correspond to negative
interactions, red arrows to positive ones. | ||
|
Inhibition of COP1 activity in light. Three pathways of ÑÎÐ1 deactivation can be distinguished. The quickest response is due to the direct interaction of photoreceptor molecules with ÑÎÐ1. As a result, the transcription of some light-activated genes is derepressed, including cip4 and cip7, and the next pathway, involving interaction of COP1 with CIP4, CIP7, and SPA1 nuclear proteins, is activated .At the final stage, COP1, which is localized in the nucleus in darkness, is transported from the nucleus to the cytoplasm, where it binds to the CIP1 and CIP8 proteins. |
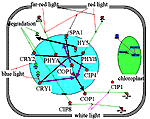
"COP1_light" filter in the Photomorphogenesis gene network. Repression of COP1 activity by light is realized through PHYB, CRY1, CRY2 photoreceptors and SPA, CIP4, CIP7 proteins. | ||
|
Light activates positive and negative regulators of photomorphogenesis. Arabidopsis COP1 is a negative regulator of photomorphogenesis, which targets HY5, a positive regulator of photomorphogenesis, for degradation via the proteasome pathway in the absence of light. The repression of COP1 in light results in a decrease in HY5 degradation. Phosphorylation affects the HY5 activity. A reduction in protein kinase CK2 activity leads to an increase in the HY5 active nonphosphorylated protein pool. The amount of active HY5 increases in light due to an increase in hy5 gene transcription. Along with positive regulators, light activates some negative regulators that decrease the intensity of the seedling response to the light signal. SUB1, a light-activated negative regulator, limits the activity of the transcription factor HY5. |
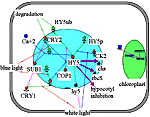
Application of "HY5" filter to Photomorphogenesis gene network. Activation of HY5 transcription factor by light induces hypocotyl growth inhibition, activation transcription of chs and rbcS genes. | ||
|
Inhibition of hypocotyl growth. Hypocotyl is a part of the stem between the root and cotyledon leaves. Low hypocotyl growth in light is a result of the balance between positively and negatively regulated cell processes. Hypocotyl grows intensely in darkness because of involvement of the COP1 repressor and PRA2 and ATHB-2 proteins. After light treatment transcription factors HY5 and HFR1 act as positive regulators, whereas proteins SPA1, EID1, PKS1, and SUB1 have negative effect on the hypocotyl growth inhibition. FHY1, a positive component of the PHYÀ signal transduction pathway, can be either a positive or a negative regulator of the PHYB signal pathway of hypocotyl growth regulation. |
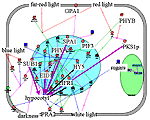
Application of "hypocotyl" filter to the Photomorphogenesis gene network. Inhibition of hypocotyl growth by light. HY5 and HFR1 inhibit hypocotyl growth while SPA1, EID1, PKS1, and SUB1 have opposite effect. | ||
|
Anthocyanin accumulation in the cell. The anthocyanin accumulates in seedlings after light treatment. CHS is a key enzyme in anthocyanin synthesis. Along with positive transcription factors HY5, CPRF2, CPRF4, CPRF1/CPRF4, MYB1, light induces accumulation of the CPRF1 factor, which is a negative regulator of the chs and cprf1gene transcription. Anionic channels play an important role in anthocyanin accumulation. Phytochrome A photoreceptor is a positive regulator and phytochrome B is a negative regulator of activation of anionic channels by cryptochrome 1. |
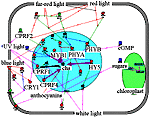
Application of "anthocyanin" filter to the Photomorphogenesis gene network. Anthocyanin accumulation in the cell. | ||
|
Formation of mature chloroplasts. RBCS and CAB proteins involve in the Calvin cycle and the formation of photosynthetic machinery. Transcription factor HY5 participates in regulation of rbcs gene. Protein kinase CK2 is inhibited in response to light, which leads to activation of rbcs and cab genes. CCA1, a cab gene transcription factor is regulated in response to light by the negative feedback principle. |
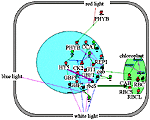 "Chloroplast" filter in the Photomorphogenesis gene network. Formation of mature chloroplasts. HY5 activates transcription of rbcS gene. | ||
| ©1997-2002, IC&G SB RAS, Laboratory of Theoretical Genetics |
Web-master Ekaterina Denisova |